Positively selected germinal center B cells (GCBC) can either resume proliferation and somatic hypermutation or differentiate. The mechanisms dictating these alternative cell fates are incompletely understood. We show that the protein arginine methyltransferase 1 (Prmt1) is upregulated in murine GCBC by Myc and mTORC-dependent signaling after positive selection. Deleting Prmt1 in activated B cells compromises antibody affinity maturation by hampering proliferation and GCBC light zone to dark zone cycling. Prmt1 deficiency also results in enhanced memory B cell generation and plasma cell differentiation, albeit the quality of these cells is compromised by the GCBC defects. We further demonstrate that Prmt1 intrinsically limits plasma cell differentiation, a function co-opted by B cell lymphoma (BCL) cells. Consistently, PRMT1 expression in BCL correlates with poor disease outcome, depends on MYC and mTORC1 activity, is required for cell proliferation, and prevents differentiation. Collectively, these data identify PRMT1 as a determinant of normal and cancerous mature B cell proliferation and differentiation balance.© 2023 Litzler et al.
In The Journal of Experimental Medicine on 4 September 2023 by Litzler, L. C., Zahn, A., et al.
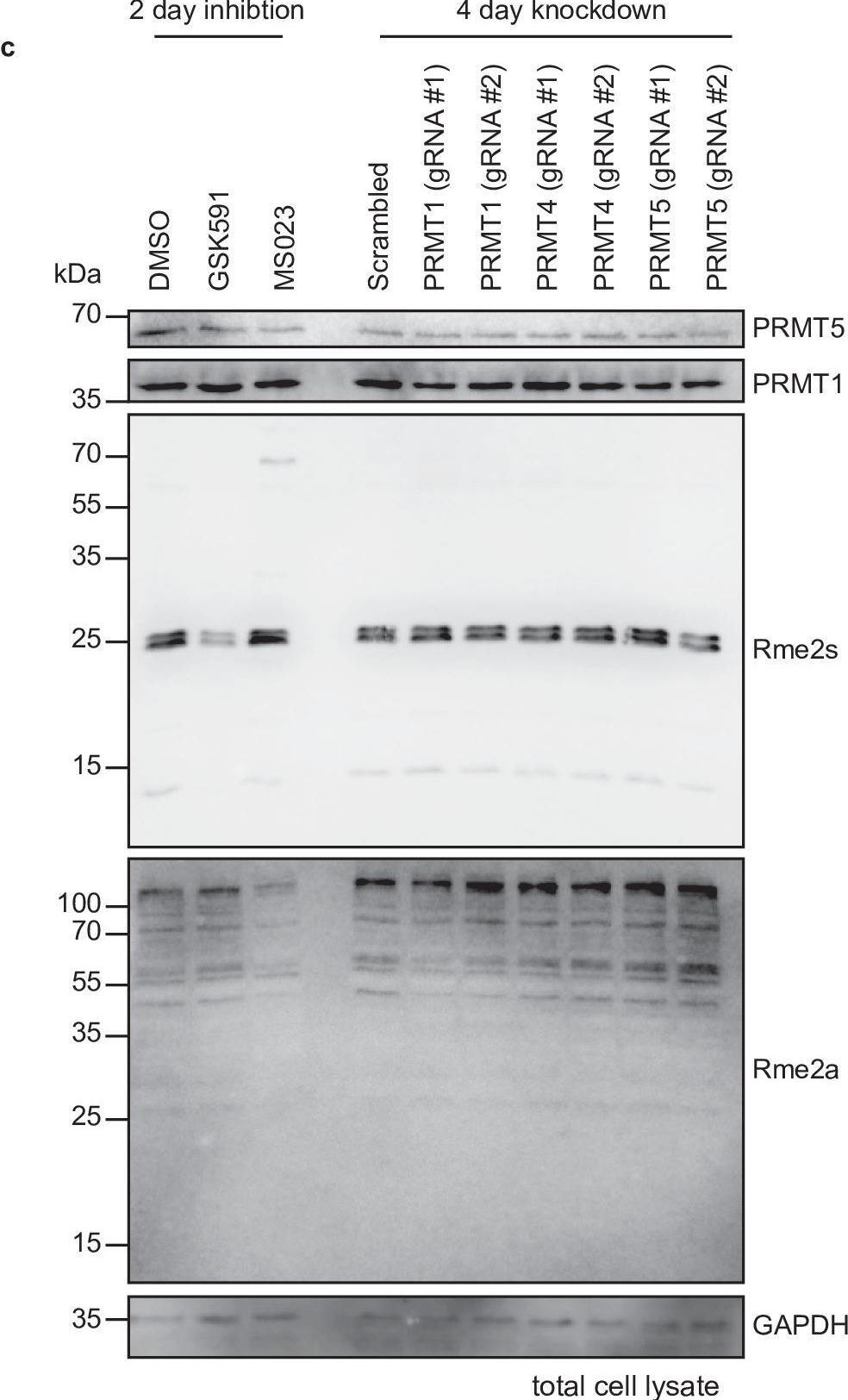
In eLife on 5 January 2022 by Maron, M. I., Casill, A. D., et al.
Protein arginine methyltransferases (PRMTs) are required for the regulation of RNA processing factors. Type I PRMT enzymes catalyze mono- and asymmetric dimethylation; Type II enzymes catalyze mono- and symmetric dimethylation. To understand the specific mechanisms of PRMT activity in splicing regulation, we inhibited Type I and II PRMTs and probed their transcriptomic consequences. Using the newly developed Splicing Kinetics and Transcript Elongation Rates by Sequencing (SKaTER-seq) method, analysis of co-transcriptional splicing demonstrated that PRMT inhibition resulted in altered splicing rates. Surprisingly, co-transcriptional splicing kinetics did not correlate with final changes in splicing of polyadenylated RNA. This was particularly true for retained introns (RI). By using actinomycin D to inhibit ongoing transcription, we determined that PRMTs post-transcriptionally regulate RI. Subsequent proteomic analysis of both PRMT-inhibited chromatin and chromatin-associated polyadenylated RNA identified altered binding of many proteins, including the Type I substrate, CHTOP, and the Type II substrate, SmB. Targeted mutagenesis of all methylarginine sites in SmD3, SmB, and SmD1 recapitulated splicing changes seen with Type II PRMT inhibition, without disrupting snRNP assembly. Similarly, mutagenesis of all methylarginine sites in CHTOP recapitulated the splicing changes seen with Type I PRMT inhibition. Examination of subcellular fractions further revealed that RI were enriched in the nucleoplasm and chromatin. Taken together, these data demonstrate that, through Sm and CHTOP arginine methylation, PRMTs regulate the post-transcriptional processing of nuclear, detained introns.
© 2022, Maron et al.
In IScience on 24 September 2021 by Maron, M. I., Lehman, S. M., et al.
Protein arginine methyltransferases (PRMTs) catalyze the post-translational monomethylation (Rme1), asymmetric (Rme2a), or symmetric (Rme2s) dimethylation of arginine. To determine the cellular consequences of type I (Rme2a) and II (Rme2s) PRMTs, we developed and integrated multiple approaches. First, we determined total cellular dimethylarginine levels, revealing that Rme2s was ∼3% of total Rme2 and that this percentage was dependent upon cell type and PRMT inhibition status. Second, we quantitatively characterized in vitro substrates of the major enzymes and expanded upon PRMT substrate recognition motifs. We also compiled our data with publicly available methylarginine-modified residues into a comprehensive database. Third, we inhibited type I and II PRMTs and performed proteomic and transcriptomic analyses to reveal their phenotypic consequences. These experiments revealed both overlapping and independent PRMT substrates and cellular functions. Overall, this study expands upon PRMT substrate diversity, the arginine methylome, and the complex interplay of type I and II PRMTs.
© 2021 The Author(s).
In FEBS Letters on 1 September 2020 by Park, M. J., Liao, J., et al.
We previously reported the involvement of protein arginine methyltransferase 1 (PRMT1) in adipocyte thermogenesis. Here, we investigate the effects of PRMT1 inhibitors on thermogenesis. Unexpectedly, we find that the PRMT1 inhibitor TC-E 5003 (TC-E) induces the thermogenic properties of primary murine and human subcutaneous adipocytes. TC-E treatment upregulates the expression of Ucp1 and Fgf21 significantly and activates protein kinase A signaling and lipolysis in primary subcutaneous adipocytes from both mouse and humans. We further find that the thermogenic effects of TC-E are independent of PRMT1 and beta-adrenergic receptors. Our data indicate that TC-E exerts strong effects on murine and human subcutaneous adipocytes by activating beige adipocytes via PKA signaling.
© 2020 Federation of European Biochemical Societies.
GFI1 facilitates efficient DNA repair by regulating PRMT1 dependent methylation of MRE11 and 53BP1.
In Nature Communications on 12 April 2018 by Vadnais, C., Chen, R., et al.
GFI1 is a transcriptional regulator expressed in lymphoid cells, and an "oncorequisite" factor required for development and maintenance of T-lymphoid leukemia. GFI1 deletion causes hypersensitivity to ionizing radiation, for which the molecular mechanism remains unknown. Here, we demonstrate that GFI1 is required in T cells for the regulation of key DNA damage signaling and repair proteins. Specifically, GFI1 interacts with the arginine methyltransferase PRMT1 and its substrates MRE11 and 53BP1. We demonstrate that GFI1 enables PRMT1 to bind and methylate MRE11 and 53BP1, which is necessary for their function in the DNA damage response. Thus, our results provide evidence that GFI1 can adopt non-transcriptional roles, mediating the post-translational modification of proteins involved in DNA repair. These findings have direct implications for treatment responses in tumors overexpressing GFI1 and suggest that GFI1's activity may be a therapeutic target in these malignancies.
In Elife on 5 January 2022 by Maron, M. I., Casill, A. D., et al.
Fig.1.C
-
WB
-
Collected and cropped from eLife by CiteAb, provided under a CC-BY license
Image 1 of 1
