Natural products ranging from phytochemicals to metals are well-known for their therapeutic benefits on different cancer types, including acute leukemia. However, bioavailability significantly limited the applications of various polyphenolic molecules, such as curcumin, while toxicity challenged the medicinal applications of heavy metals, such as mercury (Hg). Specifically, in case of curcumin derivatives, simultaneous solubility, stability, and bioactivity in the aqueous medium remain unachieved, leading to poor clinical translation. We demonstrate for the first time that the above-mentioned challenges could be resolved by covalently bonding mercury to the α-carbon of curcumin. The resultant organomercury compound ((1E,6E)-1,7-bis(4-hydroxy-3-methoxyphenyl)-3,5-dioxohepta-1,6-dien-4-yl)mercury or α-Mercurin is soluble in alkaline conditions and remains stable for at least 24 h. Cell viability assays demonstrated selective cytotoxicity of α-Mercurin against acute leukemia cells, compared to healthy human peripheral blood mononuclear cells, in vitro. Experimental IC50 on MOLT-4 and HL-60 cells remained in the lower micromolar range, and potential mode of action includes apoptosis. Ex vivo analysis also demonstrated that α-Mercurin can eliminate immature blasts from acute lymphoblastic leukemia patients' blood samples and also enhance expression of immune markers, with no notable toxicity on red blood cells as well as lymphocytes. Finally, intravenous administration of α-Mercurin showed no subacute toxicity, in vivo.
© 2025 The Authors. Published by American Chemical Society.
Product Citations: 39
In ACS Omega on 6 May 2025 by Mondal, S., Das, U., et al.
-
Cancer Research
S100A9 and HMGB1 orchestrate MDSC-mediated immunosuppression in melanoma through TLR4 signaling.
In Journal for Immunotherapy of Cancer on 11 September 2024 by Ozbay Kurt, F. G., Cicortas, B. A., et al.
Immunotherapies for malignant melanoma are challenged by the resistance developed in a significant proportion of patients. Myeloid-derived suppressor cells (MDSC), with their ability to inhibit antitumor T-cell responses, are a major contributor to immunosuppression and resistance to immune checkpoint therapies in melanoma. Damage-associated molecular patterns S100A8, S100A9, and HMGB1, acting as toll like receptor 4 (TLR4) and receptor for advanced glycation endproducts (RAGE) ligands, are highly expressed in the tumor microenvironment and drive MDSC activation. However, the role of TLR4 and RAGE signaling in the acquisition of MDSC immunosuppressive properties remains to be better defined. Our study investigates how the signaling via TLR4 and RAGE as well as their ligands S100A9 and HMGB1, shape MDSC-mediated immunosuppression in melanoma.
MDSC were isolated from the peripheral blood of patients with advanced melanoma or generated in vitro from healthy donor-derived monocytes. Monocytes were treated with S100A9 or HMGB1 for 72 hours. The immunosuppressive capacity of treated monocytes was assessed in the inhibition of T-cell proliferation assay in the presence or absence of TLR4 and RAGE inhibitors. Plasma levels of S100A8/9 and HMGB1 were quantified by ELISA. Single-cell RNA sequencing (scRNA-seq) was performed on monocytes from patients with melanoma and healthy donors.
We showed that exposure to S100A9 and HMGB1 converted healthy donor-derived monocytes into MDSC through TLR4 signaling. Our scRNA-seq data revealed in patient monocytes enriched inflammatory genes, including S100 and those involved in NF-κB and TLR4 signaling, and a reduced major histocompatibility complex II gene expression. Furthermore, elevated plasma S100A8/9 levels correlated with shorter progression-free survival in patients with melanoma.
These findings highlight the critical role of TLR4 and, to a lesser extent, RAGE signaling in the conversion of monocytes into MDSC-like cells, underscore the potential of targeting S100A9 to prevent this conversion, and highlight the prognostic value of S100A8/9 as a plasma biomarker in melanoma.
© Author(s) (or their employer(s)) 2024. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
-
Homo sapiens (Human)
-
Cancer Research
In Cell Reports Medicine on 18 June 2024 by Montalban-Bravo, G., Thongon, N., et al.
RAS pathway mutations, which are present in 30% of patients with chronic myelomonocytic leukemia (CMML) at diagnosis, confer a high risk of resistance to and progression after hypomethylating agent (HMA) therapy, the current standard of care for the disease. Here, using single-cell, multi-omics technologies, we seek to dissect the biological mechanisms underlying the initiation and progression of RAS pathway-mutated CMML. We identify that RAS pathway mutations induce transcriptional reprogramming of hematopoietic stem and progenitor cells (HSPCs) and downstream monocytic populations in response to cell-intrinsic and -extrinsic inflammatory signaling that also impair the functions of immune cells. HSPCs expand at disease progression after therapy with HMA or the BCL2 inhibitor venetoclax and rely on the NF-κB pathway effector MCL1 to maintain survival. Our study has implications for the development of therapies to improve the survival of patients with RAS pathway-mutated CMML.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
-
Homo sapiens (Human)
-
Cancer Research
In Applied Biochemistry and Biotechnology on 1 September 2023 by Sanju, S., Jain, P., et al.
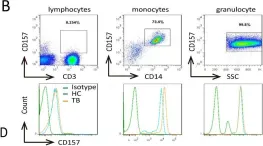
Quantitation of mHLA-DR and nCD64 is useful in understanding the dysregulated host response. The down regulation of HLA-DR expression on the circulating monocytes (mHLA-DR) is associated with anti-inflammatory response, and an increased expression of CD64 on neutrophil surface (nCD64) is associated with pro-inflammatory response. Quantitation of these antigen expression using beads (QuantiBRITE™ PE) is a precision technique. These beads are reported to be stable for 24 h after reconstitution. We report the results of our investigation examining the stability of QuantiBRITE PE beads over a period of 4-week post-reconstitution. The data suggest that reconstituted QuantiBRITE PE beads, if stored in dark at 2-8 °C, can be effectively used for up to 2 weeks for determining nCD64 and mHLA-DR antibody bound per cell (ABC) values.
© 2022. The Author(s), under exclusive licence to Springer Science+Business Media, LLC, part of Springer Nature.
-
FC/FACS
-
Homo sapiens (Human)
-
Biochemistry and Molecular biology
In Blood Cancer Discovery on 1 May 2023 by Vujovic, A., de Rooij, L. P. M. H., et al.
Acute myeloid leukemia (AML) is fueled by leukemic stem cells (LSC) whose determinants are challenging to discern from hematopoietic stem cells (HSC) or uncover by approaches focused on general cell properties. We have identified a set of RNA-binding proteins (RBP) selectively enriched in human AML LSCs. Using an in vivo two-step CRISPR-Cas9 screen to assay stem cell functionality, we found 32 RBPs essential for LSCs in MLL-AF9;NrasG12D AML. Loss-of-function approaches targeting key hit RBP ELAVL1 compromised LSC-driven in vivo leukemic reconstitution, and selectively depleted primitive malignant versus healthy cells. Integrative multiomics revealed differentiation, splicing, and mitochondrial metabolism as key features defining the leukemic ELAVL1-mRNA interactome with mitochondrial import protein, TOMM34, being a direct ELAVL1-stabilized target whose repression impairs AML propagation. Altogether, using a stem cell-adapted in vivo CRISPR screen, this work demonstrates pervasive reliance on RBPs as regulators of LSCs and highlights their potential as therapeutic targets in AML.
LSC-targeted therapies remain a significant unmet need in AML. We developed a stem-cell-adapted in vivo CRISPR screen to identify key LSC drivers. We uncover widespread RNA-binding protein dependencies in LSCs, including ELAVL1, which we identify as a novel therapeutic vulnerability through its regulation of mitochondrial metabolism. This article is highlighted in the In This Issue feature, p. 171.
©2023 The Authors; Published by the American Association for Cancer Research.
-
Genetics
-
Stem Cells and Developmental Biology
In MBio on 27 August 2019 by Yang, Q., Liao, M., et al.
Fig.1.B

-
FC/FACS
-
Collected and cropped from MBio by CiteAb, provided under a CC-BY license
Image 1 of 1