Homologous recombination (HR) is crucial for the high-fidelity repair of DNA double-strand breaks (DSBs), ensuring the maintenance of genome stability. In this study, we show that FOXD3 interacts with poly (ADP-ribose) polymerase 1 (PARP1) and is recruited to DSBs in a PARP1-dependent manner. FOXD3 directly binds to the DSB repair protein MRE11 and promotes its recruitment to DSB sites, ensuring proper end resection. Inhibition of FOXD3 expression compromises HR-mediated DSB repair and chromosome stability and sensitizes cancer cells to ionizing radiation. Collectively, our findings demonstrate that FOXD3 promotes HR-mediated DSB repair and genome stability.
Product Citations: 1,423
In Acta Biochimica et Biophysica Sinica on 13 June 2025 by Xu, S., Zhang, J., et al.
-
Genetics
The class II myosin MYH4 safeguards genome integrity and suppresses tumor progression.
In The Journal of Clinical Investigation on 2 June 2025 by Thatte, J., Moisés da Silva, A., et al.
Loss-of-function mutations in genome maintenance genes fuel tumorigenesis through increased genomic instability. A subset of these tumor suppressors are challenging to identify due to context dependency, including functional interactions with other genes and pathways. Here, we searched for potential causal genes that impact tumor development and/or progression in breast cancer through functional-genetic screening of candidate genes. MYH4, encoding a class II myosin, emerged as a top hit impacting genomic stability. We show that MYH4 suppresses DNA replication stress by promoting replication licensing and replication fork progression. Moreover, we observed a strong synergistic relationship among class II myosins in suppressing replication-associated DNA damage. Genomic analysis of Pan-Cancer Analysis of Whole Genomes project breast cancer samples revealed frequent concomitant loss of TP53 with MYH4 and class II myosins on chromosome 17p. Notably, Myh4 disruption accelerated mouse mammary tumorigenesis in a Trp53-deficient background. In conclusion, our results suggest an unanticipated function of MYH4 in p53-mediated tumor suppression that can explain their combined loss in breast cancer.
-
Cancer Research
Multigenerational cell tracking of DNA replication and heritable DNA damage.
In Nature on 1 June 2025 by Panagopoulos, A., Stout, M., et al.
Cell heterogeneity is a universal feature of life. Although biological processes affected by cell-to-cell variation are manifold, from developmental plasticity to tumour heterogeneity and differential drug responses, the sources of cell heterogeneity remain largely unclear1,2. Mutational and epigenetic signatures from cancer (epi)genomics are powerful for deducing processes that shaped cancer genome evolution3-5. However, retrospective analyses face difficulties in resolving how cellular heterogeneity emerges and is propagated to subsequent cell generations. Here, we used multigenerational single-cell tracking based on endogenously labelled proteins and custom-designed computational tools to elucidate how oncogenic perturbations induce sister cell asymmetry and phenotypic heterogeneity. Dual CRISPR-based genome editing enabled simultaneous tracking of DNA replication patterns and heritable endogenous DNA lesions. Cell lineage trees of up to four generations were tracked in asynchronously growing cells, and time-resolved lineage analyses were combined with end-point measurements of cell cycle and DNA damage markers through iterative staining. Besides revealing replication and repair dynamics, damage inheritance and emergence of sister cell heterogeneity across multiple cell generations, through combination with single-cell transcriptomics, we delineate how common oncogenic events trigger multiple routes towards polyploidization with distinct outcomes for genome integrity. Our study provides a framework to dissect phenotypic plasticity at the single-cell level and sheds light onto cellular processes that may resemble early events during cancer development.
© 2025. The Author(s).
-
Genetics
In Cell Death and Differentiation on 29 May 2025 by Modafferi, S., Farina, S., et al.
Formation of cytoplasmic inclusions (CIs) of TDP-43 and FUS, along with DNA damage accumulation, is a hallmark of affected motor neurons in Amyotrophic Lateral Sclerosis (ALS). However, the impact of CIs on DNA damage response (DDR) and repair in this pathology remains unprobed. Here, we show that CIs of TDP-43 and FUSP525L, co-localizing with stress granules, lead to a dysfunctional DDR activation associated with physical DNA breakage. Inhibition of the activity of the DDR kinase ATM, but not of ATR, abolishes DDR signaling, indicating that DNA double-strand breaks (DSBs) are the primary source of DDR activation. In addition, cells with TDP-43 and FUSP525L CIs exhibit reduced DNA damage-induced RNA synthesis at DSBs. We previously showed that the two endoribonucleases DROSHA and DICER, also known to interact with TDP-43 and FUS during small RNA processing, contribute to DDR signaling at DSBs. Treatment with enoxacin, which stimulates DDR and repair by boosting the enzymatic activity of DICER, restores a proficient DDR and reduces DNA damage accumulation in cultured cells with CIs and in vivo in a murine model of ALS. In Drosophila melanogaster, Dicer-2 overexpression rescues TDP-43-mediated retinal degeneration. In summary, our results indicate that the harmful effects caused by TDP-43 and FUS CIs include genotoxic stress and that the pharmacological stimulation of the DNA damage signaling and repair counteracts it.
© 2025. The Author(s).
-
Cell Biology
-
Genetics
In Zoological Research on 18 May 2025 by Huang, X. Y., Liu, X. Y., et al.
The DNA replication stress (RS) response is crucial for maintaining cellular homeostasis and promoting physiological longevity. However, the mechanisms by which long-lived species, such as bats, regulate RS to maintain genomic stability remain unclear. Also, recent studies have uncovered noncanonical roles of ribosome-associated factors in maintaining genomic stability. In this study, somatic skin fibroblasts from the long-lived big-footed bat ( Myotis pilosus) were examined, with results showing that bat cells exhibited enhanced RS tolerance compared to mouse cells. Comparative transcriptome analysis under RS conditions revealed pronounced species-specific transcriptional differences, including robust up-regulation of ribosome biogenesis genes in bat cells and a markedly reduced activation of the P53 signaling pathway. These features emphasize a distinct homeostatic strategy in bat cells. Nuclear fragile X mental retardation-interacting protein 1 ( Nufip1), a ribosome-associated factor highly expressed in bat fibroblasts, was identified as a potential integrator of ribosomal and P53 signaling via its association with ribosomal protein S27-like (Rps27l). These findings provide direct cellular and molecular evidence for a noncanonical RS response in bats, highlighting a deeper understanding of the biological characteristics and genomic maintenance mechanisms of long-lived species.
-
Genetics
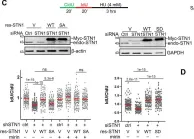
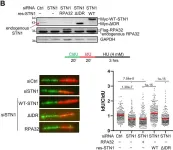
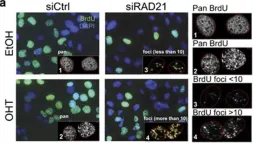
In Nat Commun on 30 November 2023 by Jaiswal, R. K., Lei, K. H., et al.
Fig.2.C

-
WB
-
Collected and cropped from Nat Commun by CiteAb, provided under a CC-BY license
Image 1 of 29
In Nat Commun on 30 November 2023 by Jaiswal, R. K., Lei, K. H., et al.
Fig.1.B

-
WB
-
Collected and cropped from Nat Commun by CiteAb, provided under a CC-BY license
Image 1 of 29
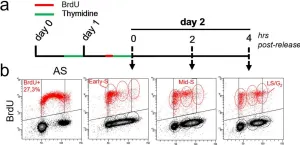
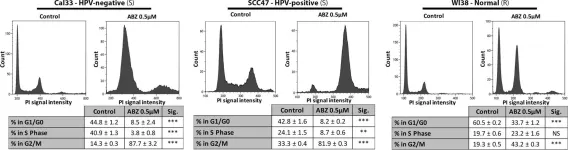
In Commun Biol on 2 March 2023 by Maresca, C., Dello Stritto, A., et al.
Fig.1.A

-
FC/FACS
-
Collected and cropped from Commun Biol by CiteAb, provided under a CC-BY license
Image 1 of 29
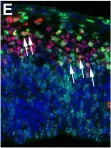
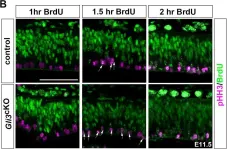
In Elife on 25 August 2020 by Hasenpusch-Theil, K., Laclef, C., et al.
Fig.3.E

-
IHC-IF
-
Mus musculus (House mouse)
Collected and cropped from Elife by CiteAb, provided under a CC-BY license
Image 1 of 29
In Elife on 25 August 2020 by Hasenpusch-Theil, K., Laclef, C., et al.
Fig.3.A

-
IHC-IF
-
Mus musculus (House mouse)
Collected and cropped from Elife by CiteAb, provided under a CC-BY license
Image 1 of 29
In Elife on 25 August 2020 by Hasenpusch-Theil, K., Laclef, C., et al.
Fig.3.B

-
IHC-IF
-
Mus musculus (House mouse)
Collected and cropped from Elife by CiteAb, provided under a CC-BY license
Image 1 of 29
In Elife on 25 August 2020 by Hasenpusch-Theil, K., Laclef, C., et al.
Fig.3.D

-
IHC-IF
-
Mus musculus (House mouse)
Collected and cropped from Elife by CiteAb, provided under a CC-BY license
Image 1 of 29
In Nat Commun on 13 December 2019 by Brison, O., El-Hilali, S., et al.
Fig.4.C

-
ICC-IF
-
Homo sapiens (Human)
Collected and cropped from Nat Commun by CiteAb, provided under a CC-BY license
Image 1 of 29
In Int J Mol Sci on 13 December 2019 by Le-Bel, G., Cortez Ghio, S., et al.
Fig.7.A

-
ICC-IF
-
Homo sapiens (Human)
Collected and cropped from Int J Mol Sci by CiteAb, provided under a CC-BY license
Image 1 of 29
In Development on 20 August 2018 by Hasenpusch-Theil, K., West, S., et al.
Fig.2.H

-
IHC-IF
-
Mus musculus (House mouse)
Collected and cropped from Development by CiteAb, provided under a CC-BY license
Image 1 of 29
In Development on 20 August 2018 by Hasenpusch-Theil, K., West, S., et al.
Fig.2.G

-
IHC
-
Mus musculus (House mouse)
Collected and cropped from Development by CiteAb, provided under a CC-BY license
Image 1 of 29
In Development on 20 August 2018 by Hasenpusch-Theil, K., West, S., et al.
Fig.4.A

-
IHC
-
Mus musculus (House mouse)
Collected and cropped from Development by CiteAb, provided under a CC-BY license
Image 1 of 29
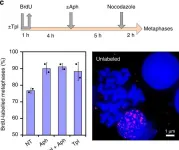
In Development on 20 August 2018 by Hasenpusch-Theil, K., West, S., et al.
Fig.4.B

-
IHC
-
Mus musculus (House mouse)
Collected and cropped from Development by CiteAb, provided under a CC-BY license
Image 1 of 29
In Oncotarget on 22 September 2017 by Ghasemi, F., Black, M., et al.
Fig.4.A

-
WB
-
Homo sapiens (Human)
Collected and cropped from Oncotarget by CiteAb, provided under a CC-BY license
Image 1 of 29
In Cell Death Dis on 27 July 2017 by Rohban, S., Cerutti, A., et al.
Fig.3.A

-
ICC-IF
-
Collected and cropped from Cell Death Dis by CiteAb, provided under a CC-BY license
Image 1 of 29
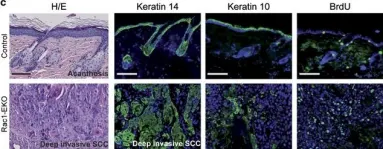
In Cell Death Dis on 9 March 2017 by Deshmukh, J., Pofahl, R., et al.
Fig.8.C

-
IHC
-
Mus musculus (House mouse)
Collected and cropped from Cell Death Dis by CiteAb, provided under a CC-BY license
Image 1 of 29
In PLoS Pathog on 1 March 2017 by Xu, P., Zhou, Z., et al.
Fig.7.A

-
FC/FACS
-
Homo sapiens (Human)
Collected and cropped from PLoS Pathog by CiteAb, provided under a CC-BY license
Image 1 of 29
In Sci Rep on 23 August 2016 by Halin Bergström, S., Hägglöf, C., et al.
Fig.3.B

-
IHC
-
Rattus norvegicus (Rat)
Collected and cropped from Sci Rep by CiteAb, provided under a CC-BY license
Image 1 of 29
In Open Biol on 1 January 2016 by Ligasová, A., Liboska, R., et al.
Fig.1.A

-
ICC-IF
-
Homo sapiens (Human)
Collected and cropped from Open Biol by CiteAb, provided under a CC-BY license
Image 1 of 29
In Open Biol on 1 January 2016 by Ligasová, A., Liboska, R., et al.
Fig.6.A

-
ICC-IF
-
Homo sapiens (Human)
Collected and cropped from Open Biol by CiteAb, provided under a CC-BY license
Image 1 of 29
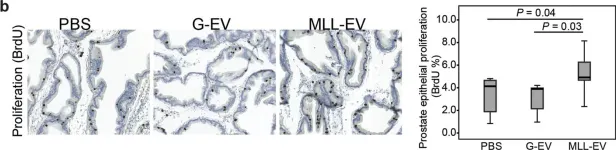
In Aging Cell on 1 February 2015 by Boquoi, A., Arora, S., et al.
Fig.2.A

-
IHC
-
Mus musculus (House mouse)
Collected and cropped from Aging Cell by CiteAb, provided under a CC-BY license
Image 1 of 29
In Aging Cell on 1 February 2015 by Boquoi, A., Arora, S., et al.
Fig.1.B

-
IHC
-
Mus musculus (House mouse)
Collected and cropped from Aging Cell by CiteAb, provided under a CC-BY license
Image 1 of 29
In Regeneration (Oxf) on 1 February 2014 by Wu, P., Alibardi, L., et al.
Fig.1.K,L,M

-
IHC-IF
-
Lizard
Collected and cropped from Regeneration (Oxf) by CiteAb, provided under a CC-BY license
Image 1 of 29
In Regeneration (Oxf) on 1 February 2014 by Wu, P., Alibardi, L., et al.
Fig.1.H,I,J

-
IHC
-
Lizard
Collected and cropped from Regeneration (Oxf) by CiteAb, provided under a CC-BY license
Image 1 of 29
In PLoS One on 12 April 2012 by Reich, J. & Papoulas, O.
Fig.5.D

-
ICC-IF
-
Mus musculus (House mouse)
Collected and cropped from PLoS One by CiteAb, provided under a CC-BY license
Image 1 of 29