Ubiquitin-specific protease 8 exerts multiple cellular functions and was identified as a potential target in a multiple myeloma vulnerability screen. Here we characterized the function of USP8 in B cells and multiple myeloma, and analyzed its impact on the global and ubiquitin-modified proteome. Usp8 deletion in mice starting at the the pre-B cell stage caused a partial block in B cell development favoring immature and innate-like B cells, as well as germinal center and plasma cells. This was accompanied by elevated immune-responses and Roquin depletion. Accordingly, correlation analyses in multiple myeloma patients revealed that low USP8 expression at diagnosis correlates with decreased survival. B cells expressing catalytically inactive USP8 accumulate protein modified with mixed ubiquitin/NEDD8 chains as hallmarks of proteotoxic stress, which we identified as favored USP8 substrates. USP8 knockdown reduced survival of bortezomib-resistant multiple myeloma cells in a lysosomal dysfunction-dependent manner. In contrast, the inhibitor DUB-IN-2 resensitized bortezomib-resistant multiple myeloma cells to treatment in a bortezomib-synergistic manner. Hence, our analyses uncovered the therapeutic potential of USP8 inhibition and of DUB-IN-2 in multiple myeloma.
Product Citations: 107
Ubiquitin-specific protease 8 controls B cell proteostasis and cell survival in multiple myeloma
Preprint on BioRxiv : the Preprint Server for Biology on 26 April 2024 by Dufner, A., Thery, F., et al.
-
Mus musculus (House mouse)
-
Immunology and Microbiology
Mutual modulation of gut microbiota and the immune system in type 1 diabetes models.
In Nature Communications on 27 November 2023 by Rosell-Mases, E., Santiago, A., et al.
The transgenic 116C-NOD mouse strain exhibits a prevalent Th17 phenotype, and reduced type 1 diabetes (T1D) compared to non-obese diabetic (NOD) mice. A cohousing experiment between both models revealed lower T1D incidence in NOD mice cohoused with 116C-NOD, associated with gut microbiota changes, reduced intestinal permeability, shifts in T and B cell subsets, and a transition from Th1 to Th17 responses. Distinct gut bacterial signatures were linked to T1D in each group. Using a RAG-2-/- genetic background, we found that T cell alterations promoted segmented filamentous bacteria proliferation in young NOD and 116C-NOD, as well as in immunodeficient NOD.RAG-2-/- and 116C-NOD.RAG-2-/- mice across all ages. Bifidobacterium colonization depended on lymphocytes and thrived in a non-diabetogenic environment. Additionally, 116C-NOD B cells in 116C-NOD.RAG-2-/- mice enriched the gut microbiota in Adlercreutzia and reduced intestinal permeability. Collectively, these results indicate reciprocal modulation between gut microbiota and the immune system in rodent T1D models.
© 2023. The Author(s).
-
Mus musculus (House mouse)
-
Immunology and Microbiology
Mutual Modulation of Gut Microbiota and the Immune System in Type 1 Diabetes Models
Preprint on Research Square on 7 February 2023 by Rosell-Mases, E., Santiago, A., et al.
Objective: Type 1 diabetes (T1D) has been associated with alterations of the gut microbiota. Here we investigate the cross-talk between the immune system and the intestinal microbiota in murine T1D. Design: To evaluate the modulation of T1D by gut microbiota, non-obese diabetic (NOD) mice were cohoused with the 116C-NOD B-cell transgenic model. To further explore the influence of the adaptive immune system of NOD and 116C-NOD models on their fecal microbiota, we studied the immunodeficient variants NOD.RAG-2 -/- and 116C-NOD.RAG-2 -/- , as well as a non-T1D-prone mouse control. The role of B and T cells in modulating the gut microbiota composition was analyzed via intravenous injection of lymphocytes. Only female mice were studied. Results: NOD cohoused with 116C-NOD exhibited a reduction of T1D incidence. This incidence decrease was associated with a shift from a Th1 to a Th17 immune response and was driven by intestinal microbiota changes. Moreover, T1D could be predicted by different gut bacterial signatures in each group of T1D-prone mice. The proliferation of segmented filamentous bacteria, known as immune modulatory organisms, was enabled by the absence of T lymphocytes in young NOD, 116C-NOD, and immunodeficient NOD.RAG-2 -/- and 116C-NOD.RAG-2 -/- at all ages. Conversely, Bifidobacterium colonization required the presence of lymphocytes and was boosted in a non-diabetogenic milieu. Finally, 116C-NOD B cells enriched the gut microbiota of 116C-NOD.RAG-2 -/- in Adlercreutzia . Conclusion: Together, these findings evidence the reciprocal modulation of gut microbiota and the immune system in rodent models of T1D.
-
Mus musculus (House mouse)
-
Immunology and Microbiology
Preprint on BioRxiv : the Preprint Server for Biology on 31 December 2022 by Catena, X., Contreras-Alcalde, M., et al.
ABSTRACT Cutaneous melanomas are a prime example of potentially immunogenic tumors, and as such, ideal targets for immune therapy. These lesions have the largest mutational burden described to date, and accumulate a broad spectrum of post-transcriptional and translational alterations that could conceptually result in a plethora of neoantigens for immune recognition. However, a significant fraction of metastatic melanoma patients is or becomes resistant to current immunotherapeutic agents. How lesions that should represent an inherently hot milieu for immune attack shift into immunologically cold or irresponsive neoplasms is not well understood. Combining cellular systems, mouse models and clinical datasets, here we identify the growth factor Midkine (MDK) as a multipronged blocker of antigen presentation. Mechanistically, we found MDK to repress all main aspects of the maturation, activation and function of dendritic cells, particularly of conventional type 1 (cDC1). These roles of MDK were found to involve primary tumors and lymph nodes, and were traced back to suppressive effects on myeloid precursor cells in the bone marrow. Moreover, MDK shifted the transcriptional profile of DCs towards a tolerogenic state that prevented and bypassed CD8 + T cell activation. Blocking MDK enhanced the response to DC-based vaccination and improved the response to immune checkpoint blockade.Together, these data provide insight into how melanomas overcome immune surveillance and support MDK as a target for therapeutic intervention.
-
Mus musculus (House mouse)
-
Cancer Research
-
Immunology and Microbiology
In Microorganisms on 22 June 2022 by Margaroni, M., Agallou, M., et al.
Leishmania parasites are capable of effectively invading dendritic cells (DCs), a cell population orchestrating immune responses against several diseases, including leishmaniasis, by bridging innate and adaptive immunity. Leishmania on the other hand has evolved various mechanisms to subvert DCs activation and establish infection. Thus, the transcriptional profile of DCs derived from bone marrow (BMDCs) that have been infected with Leishmania infantum parasite or of DCs exposed to chemically inactivated parasites was investigated via RNA sequencing, aiming to better understand the host-pathogen interplay. Flow cytometry analysis revealed that L. infantum actively inhibits maturation of not only infected but also bystander BMDCs. Analysis of double-sorted L. infantum infected BMDCs revealed significantly increased expression of genes mainly associated with metabolism and particularly glycolysis. Moreover, differentially expressed genes (DEGs) related to DC-T cell interactions were also found to be upregulated exclusively in infected BMDCs. On the contrary, transcriptome analysis of fixed parasites containing BMDCs indicated that energy production was mediated through TCA cycle and oxidative phosphorylation. In addition, DEGs related to differentiation of DCs leading to activation and differentiation of Th17 subpopulations were detected. These findings suggest an important role of metabolism on DCs-Leishmania interplay and eventually disease establishment.
-
Biochemistry and Molecular biology
-
Immunology and Microbiology
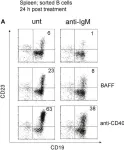
In Front Immunol on 22 August 2018 by Hobeika, E., Dautzenberg, M., et al.
Fig.6.A

-
FC/FACS
-
Collected and cropped from Front Immunol by CiteAb, provided under a CC-BY license
Image 1 of 3
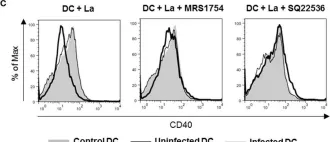
In Front Immunol on 10 August 2017 by Figueiredo, A. B., Souza-Testasicca, M. C., et al.
Fig.1.C

-
FC/FACS
-
Collected and cropped from Front Immunol by CiteAb, provided under a CC-BY license
Image 1 of 3
In Mol Cancer on 9 November 2005 by Han, S. S., Shaffer, A. L., et al.
Fig.1.B
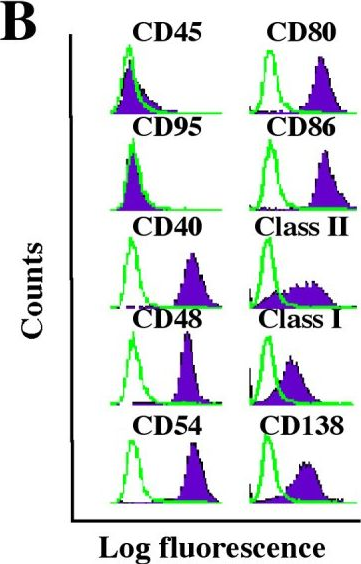
-
FC/FACS
-
Mus musculus (House mouse)
Collected and cropped from Mol Cancer by CiteAb, provided under a CC-BY license
Image 1 of 3