The immune evasion of emerging SARS-CoV-2 variants significantly undermines current vaccination efforts, calling for an updated vaccine composition. To identify optimal booster candidates against circulating JN.1, a panel of variant spikes were characterized. The omicron spikes exhibited reduced plasma membrane expression, accompanied by lower cell-cell fusion but increased viral entry. Regimens with DNA prime-DNA boost or DNA prime-adenoviral vectored vaccine boost by intramuscular immunization elicited neutralizing antibody (NAbs) and T cell responses against all variants except BA.2.86 and JN.1. Intranasal immunization induced high IgA and NAb titers in bronchoalveolar lavage against all variants except BA.2.86 and JN.1. T cell responses were generally comparable for all immunogens tested. JN.1 completely escaped NAbs in one immunized cohort, and breakthrough infections marginally boosted antibody titers. Overall, this study indicates intrinsic difficulty in eliciting NAbs against the JN.1 strain, whereas vaccines based on XBB and EG.5.1 are relatively superior in generating cross-reactive NAbs.
© 2024 The Authors.
Product Citations: 85
In IScience on 16 August 2024 by Fan, J., Zhang, Y., et al.
In Nature Communications on 14 August 2024 by Münchhalfen, M., Goerg, R., et al.
Ligation of the B cell antigen receptor (BCR) initiates humoral immunity. However, BCR signaling without appropriate co-stimulation commits B cells to death rather than to differentiation into immune effector cells. How BCR activation depletes potentially autoreactive B cells while simultaneously primes for receiving rescue and differentiation signals from cognate T lymphocytes remains unknown. Here, we use a mass spectrometry-based proteomic approach to identify cytosolic/nuclear shuttling elements and uncover transcription factor EB (TFEB) as a central BCR-controlled rheostat that drives activation-induced apoptosis, and concurrently promotes the reception of co-stimulatory rescue signals by supporting B cell migration and antigen presentation. CD40 co-stimulation prevents TFEB-driven cell death, while enhancing and prolonging TFEB's nuclear residency, which hallmarks antigenic experience also of memory B cells. In mice, TFEB shapes the transcriptional landscape of germinal center B cells. Within the germinal center, TFEB facilitates the dark zone entry of light-zone-residing centrocytes through regulation of chemokine receptors and, by balancing the expression of Bcl-2/BH3-only family members, integrates antigen-induced apoptosis with T cell-provided CD40 survival signals. Thus, TFEB reprograms antigen-primed germinal center B cells for cell fate decisions.
© 2024. The Author(s).
-
FC/FACS
-
Homo sapiens (Human)
-
Mus musculus (House mouse)
-
Immunology and Microbiology
The ATP-exporting channel Pannexin 1 promotes CD8+ T cell effector and memory responses.
In IScience on 19 July 2024 by Vardam-Kaur, T., Banuelos, A., et al.
Sensing of extracellular ATP (eATP) controls CD8+ T cell function. Their accumulation can occur through export by specialized molecules, such as the release channel Pannexin 1 (Panx1). Whether Panx1 controls CD8+ T cell immune responses in vivo, however, has not been previously addressed. Here, we report that T-cell-specific Panx1 is needed for CD8+ T cell responses to viral infections and cancer. We found that CD8-specific Panx1 promotes both effector and memory CD8+ T cell responses. Panx1 favors initial effector CD8+ T cell activation through extracellular ATP (eATP) export and subsequent P2RX4 activation, which helps promote full effector differentiation through extracellular lactate accumulation and its subsequent recycling. In contrast, Panx1 promotes memory CD8+ T cell survival primarily through ATP export and subsequent P2RX7 engagement, leading to improved mitochondrial metabolism. In summary, Panx1-mediated eATP export regulates effector and memory CD8+ T cells through distinct purinergic receptors and different metabolic and signaling pathways.
© 2024 The Author(s).
-
Immunology and Microbiology
In Front Allergy on 7 June 2024 by Walker, M. T., Bloodworth, J. C., et al.
Previous research showed that 5-hydroxytryptophan (5HTP), a metabolic precursor of serotonin, reduces allergic lung inflammation by inhibiting eosinophil migration across endothelial monolayers.
It is unknown if serotonin receptors are involved in mediating this 5HTP function or if serotonin receptor (HTR) single nucleotide polymorphisms (SNPs) associate with lung function in humans.
Serotonin receptor subtypes were assessed by qPCR, western blot, confocal microscopy, pharmacological inhibitors and siRNA knockdown. HTR SNPs were assessed in two cohorts.
Pharmacological inhibition or siRNA knockdown of the serotonin receptors HTR1A or HTR1B in endothelial cells abrogated the inhibitory effects of 5HTP on eosinophil transendothelial migration. In contrast, eosinophil transendothelial migration was not inhibited by siRNA knockdown of HTR1A or HTR1B in eosinophils. Surprisingly, these HTRs were intracellular in endothelial cells and an extracellular supplementation with serotonin did not inhibit eosinophil transendothelial migration. This is consistent with the inability of serotonin to cross membranes, the lack of selective serotonin reuptake receptors on endothelial cells, and the studies showing minimal impact of selective serotonin reuptake inhibitors on asthma. To extend our HTR studies to humans with asthma, we examined the CHIRAH and GALA cohorts for HTR SNPs that affect HTR function or are associated with behavior disorders. A polygenic index of SNPs in HTRs was associated with lower lung function in asthmatics.
Serotonin receptors mediate 5HTP inhibition of transendothelial migration and HTR SNPs associate with lower lung function. These results may serve to aid in design of novel interventions for allergic inflammation.
© 2024 Walker, Bloodworth, Kountz, McCarty, Green, Ferrie, Campbell, Averill, Beckman, Grammer, Eng, Avila, Farber, Rodriguez-Cintron, Rodriguez-Santana, Serebrisky, Thyne, Seibold, Burchard, Kumar and Cook-Mills.
Machine-learning and mechanistic modeling of metastatic breast cancer after neoadjuvant treatment.
In PLoS Computational Biology on 1 May 2024 by Benzekry, S., Mastri, M., et al.
Clinical trials involving systemic neoadjuvant treatments in breast cancer aim to shrink tumors before surgery while simultaneously allowing for controlled evaluation of biomarkers, toxicity, and suppression of distant (occult) metastatic disease. Yet neoadjuvant clinical trials are rarely preceded by preclinical testing involving neoadjuvant treatment, surgery, and post-surgery monitoring of the disease. Here we used a mouse model of spontaneous metastasis occurring after surgical removal of orthotopically implanted primary tumors to develop a predictive mathematical model of neoadjuvant treatment response to sunitinib, a receptor tyrosine kinase inhibitor (RTKI). Treatment outcomes were used to validate a novel mathematical kinetics-pharmacodynamics model predictive of perioperative disease progression. Longitudinal measurements of presurgical primary tumor size and postsurgical metastatic burden were compiled using 128 mice receiving variable neoadjuvant treatment doses and schedules (released publicly at https://zenodo.org/records/10607753). A non-linear mixed-effects modeling approach quantified inter-animal variabilities in metastatic dynamics and survival, and machine-learning algorithms were applied to investigate the significance of several biomarkers at resection as predictors of individual kinetics. Biomarkers included circulating tumor- and immune-based cells (circulating tumor cells and myeloid-derived suppressor cells) as well as immunohistochemical tumor proteins (CD31 and Ki67). Our computational simulations show that neoadjuvant RTKI treatment inhibits primary tumor growth but has little efficacy in preventing (micro)-metastatic disease progression after surgery and treatment cessation. Machine learning algorithms that included support vector machines, random forests, and artificial neural networks, confirmed a lack of definitive biomarkers, which shows the value of preclinical modeling studies to identify potential failures that should be avoided clinically.
Copyright: © 2024 Benzekry et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
-
Cancer Research
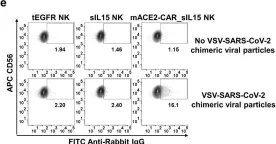
In Nat Commun on 11 May 2022 by Lu, T., Ma, R., et al.
Fig.2.E

-
FC/FACS
-
Collected and cropped from Nat Commun by CiteAb, provided under a CC-BY license
Image 1 of 2
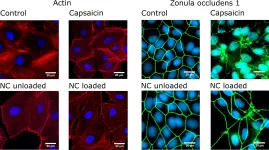
In PLoS One on 7 November 2017 by Kaiser, M., Pohl, L., et al.
Fig.5.A

-
ICC-IF
-
Canis lupus familiaris (Domestic dog)
Collected and cropped from PLoS One by CiteAb, provided under a CC-BY license
Image 1 of 2