Endometrial carcinoma (EMC) is associated with obesity; however, the underlying mechanisms have not yet been elucidated. Peroxisome proliferator-activated receptor alpha (PPARα) is a nuclear receptor that is involved in lipid, glucose, and energy metabolism. PPARα reportedly functions as a tumor suppressor through its effects on lipid metabolism; however, the involvement of PPARα in the development of EMC remains unclear. The present study demonstrated that the immunohistochemical expression of nuclear PPARα was lower in EMC than in normal endometrial tissues, suggesting the tumor suppressive nature of PPARα. A treatment with the PPARα activator, irbesartan, inhibited the EMC cell lines, Ishikawa and HEC1A, by down-regulating sterol regulatory element-binding protein 1 (SREBP1) and fatty acid synthase (FAS) and up-regulating the tumor suppressor genes p21 and p27, antioxidant enzymes, and AT-rich interaction domain 1A (ARID1A). These results indicate the potential of the activation of PPARα as a novel therapeutic approach against EMC.
© 2023 Lu et al.
Product Citations: 46
In Oncology Research on 12 June 2023 by Lu, Y. U., Miyamoto, T., et al.
-
WB
-
Homo sapiens (Human)
-
Cancer Research
PRPF19 regulates p53-dependent cellular senescence by modulating alternative splicing of MDM4 mRNA.
In The Journal of Biological Chemistry on 1 July 2021 by Yano, K., Takahashi, R. U., et al.
Alteration of RNA splicing is a hallmark of cellular senescence, which is associated with age-related disease and cancer development. However, the roles of splicing factors in cellular senescence are not fully understood. In this study, we identified the splicing factor PRPF19 as a critical regulator of cellular senescence in normal human diploid fibroblasts. PRPF19 was downregulated during replicative senescence, and PRPF19 knockdown prematurely induced senescence-like cell cycle arrest through the p53-p21 pathway. RNA-sequencing analysis revealed that PRPF19 knockdown caused a switch of the MDM4 splicing isoform from stable full-length MDM4-FL to unstable MDM4-S lacking exon 6. We also found that PRPF19 regulates MDM4 splicing by promoting the physical interaction of other splicing factors, PRPF3 and PRPF8, which are key components of the core spliceosome, U4/U6.U5 tri-snRNP. Given that MDM4 is a major negative regulator of p53, our findings imply that PRPF19 downregulation inhibits MDM4-mediated p53 inactivation, resulting in induction of cellular senescence. Thus, PRPF19 plays an important role in the induction of p53-dependent cellular senescence.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
-
WB
-
Biochemistry and Molecular biology
-
Genetics
Disruption of nucleocytoplasmic trafficking as a cellular senescence driver.
In Experimental & Molecular Medicine on 1 June 2021 by Park, J. H., Ryu, S. J., et al.
Senescent cells exhibit a reduced response to intrinsic and extrinsic stimuli. This diminished reaction may be explained by the disrupted transmission of nuclear signals. However, this hypothesis requires more evidence before it can be accepted as a mechanism of cellular senescence. A proteomic analysis of the cytoplasmic and nuclear fractions obtained from young and senescent cells revealed disruption of nucleocytoplasmic trafficking (NCT) as an essential feature of replicative senescence (RS) at the global level. Blocking NCT either chemically or genetically induced the acquisition of an RS-like senescence phenotype, named nuclear barrier-induced senescence (NBIS). A transcriptome analysis revealed that, among various types of cellular senescence, NBIS exhibited a gene expression pattern most similar to that of RS. Core proteomic and transcriptomic patterns common to both RS and NBIS included upregulation of the endocytosis-lysosome network and downregulation of NCT in senescent cells, patterns also observed in an aging yeast model. These results imply coordinated aging-dependent reduction in the transmission of extrinsic signals to the nucleus and in the nucleus-to-cytoplasm supply of proteins/RNAs. We further showed that the aging-associated decrease in Sp1 transcription factor expression was critical for the downregulation of NCT. Our results suggest that NBIS is a modality of cellular senescence that may represent the nature of physiological aging in eukaryotes.
-
Biochemistry and Molecular biology
In Oncology Letters on 1 January 2020 by Perrard, J., Morel, A., et al.
HPV16 is the most carcinogenic human papillomavirus and causes >50% of cervical cancers, the majority of anal cancers and 30% of oropharyngeal squamous cell carcinomas. HPV carcinogenesis relies on the continuous expression of the two main viral oncoproteins E6 and E7 that target >150 cellular proteins. Among them, epigenetic modifiers, including DNA Methyl Transferases (DNMT), are dysregulated, promoting an aberrant methylation pattern in HPV-positive cancer cells. It has been previously reported that the treatment of HPV-positive cervical cancer cells with DNMT inhibitor 5-aza-2'-deoxycytidine (5azadC) caused the downregulation of E6 expression due to mRNA destabilization that was mediated by miR-375. Recently, the T-box transcription factor 2 (TBX2) has been demonstrated to repress HPV LCR activity. In the current study, the role of TBX2 in E6 repression was investigated in HPV16 cervical cancer cell lines following 5azadC treatment. A decrease of E6 expression was accompanied by p53 and p21 restoration. While TBX2 mRNA was upregulated in 5azadC-treated SiHa and Ca Ski cells, TBX2 protein was not detectable. Furthermore, the overexpression of TBX2 protein in cervical cancer cells did not allow the repression of E6 expression. The TBX2 transcription factor is therefore unlikely to be associated with the repression of E6 following 5azadC treatment of SiHa and Ca Ski cells.
Copyright © 2020, Spandidos Publications.
-
WB
-
Cancer Research
-
Genetics
Preprint on BioRxiv : the Preprint Server for Biology on 9 July 2019 by Urisman, A., Yuan, T. L., et al.
h4>Background/h4> KRAS mutations are present in up to 30% of lung adenocarcinoma cases and are associated with poor survival. No effective targeted therapy against KRAS is currently available, and novel strategies to counteract oncogenic KRAS signaling are needed. h4>Results/h4> We used targeted proteomics to monitor abundance and site-specific phosphorylation in a network of over 150 upstream and downstream effectors of KRAS signaling in H358 cells (KRAS G12C). We compared patterns of protein regulation following sustained signaling blockade in the RAS/ERK module at two different levels, KRAS and MEK. Network-based analysis demonstrated complex non-linear patterns of regulation with wide-spread crosstalk among diverse subnetworks. Among 85 most regulated proteins in the network, only 12 proteins showed concordant regulation in response to signaling blockade at both KRAS and MEK levels, while the remainder were either specifically regulated in response to KRAS knockdown or MEK inhibition or showed orthogonal regulation in both conditions. Dephosphorylation of DNA methyltransferase 1 (DNMT1) at S714 was identified among the changes unique to KRAS knockdown, and here we elucidate the role of this phosphorylation in KRAS-dependent transcriptional silencing of tumor suppressor genes. h4>Conclusions/h4> Network-based analysis of the Ras signaling has shown complex non-linear patterns of regulation with wide-spread crosstalk among diverse subnetworks. Our work illustrates a targeted proteomics approach to functional interrogation of complex signaling networks focused on identification of readily testable hypotheses. These methods are widely applicable to diverse questions in tumor biology and other signaling paradigms.
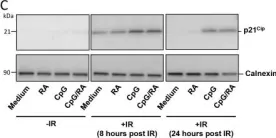
In PLoS One on 4 October 2017 by Holm, K. L., Syljuåsen, R. G., et al.
Fig.2.C

-
WB
-
Homo sapiens (Human)
Collected and cropped from PLoS One by CiteAb, provided under a CC-BY license
Image 1 of 3
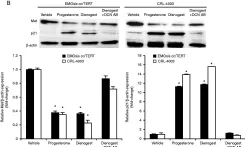
In J Endocrinol on 1 November 2014 by Ono, Y. J., Terai, Y., et al.
Fig.4.B

-
WB
-
Collected and cropped from J Endocrinol by CiteAb, provided under a CC-BY license
Image 1 of 3
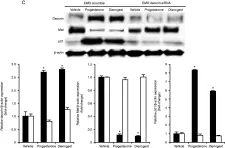
In J Endocrinol on 1 November 2014 by Ono, Y. J., Terai, Y., et al.
Fig.4.C

-
WB
-
Collected and cropped from J Endocrinol by CiteAb, provided under a CC-BY license
Image 1 of 3