The combined use of serum and CSF biomarkers for prognostic stratification in multiple sclerosis (MS) remains underexplored. This multicenter observational study investigated associations between serum neurofilament light chain (sNfL), glial fibrillary acidic protein (sGFAP), and CSF lipid-specific IgM oligoclonal bands (LS-OCMB) with different forms of disability worsening, such as relapse-associated worsening (RAW), active progression independent of relapse activity (aPIRA), and non-active PIRA (naPIRA). A total of 535 patients with MS were included, all sampled within one year of disease onset. Biomarkers were quantified using single-molecule array and immunoblotting techniques, and CSF cell subsets were analyzed by flow cytometry. High sNfL z-scores and LS-OCMB positivity were independently associated with increased risk of RAW and aPIRA, collectively termed inflammatory-associated worsening (IAW), while elevated sGFAP levels predicted naPIRA. Patients with both high sNfL and LS-OCMB positivity had the highest risk of IAW. Among LS-OCMB-positive patients, higher regulatory T cell percentages were associated with lower sNfL levels, suggesting a protective role. Conversely, in LS-OCMB-negative patients, sNfL levels correlated with CSF C3 concentrations. These findings support the complementary role of sNfL, sGFAP, and LS-OCMB in identifying distinct mechanisms of disease worsening and may inform early personalized management strategies in MS.
Product Citations: 218
In International Journal of Molecular Sciences on 18 July 2025 by Monreal, E., Fernández-Velasco, J. I., et al.
ZFP36-family RNA-binding proteins in regulatory T cells reinforce immune homeostasis.
In Nature Communications on 6 May 2025 by Sáenz-Narciso, B., Bell, S. E., et al.
RNA binding proteins (RBP) of the ZFP36 family limit the differentiation and effector functions of CD4 and CD8 T cells, but little is known of their expression or function in regulatory T (Treg) cells. By using Treg cell-restricted deletion of Zfp36 family members we identify the role of Zfp36l1 and Zfp36l2 in Treg cells to maintain immune homeostasis. Mice with Treg cells deficient in these RBP display an inflammatory phenotype with an expansion in the numbers of type-2 conventional dendritic cells, T effector cells, T follicular helper and germinal center B cells and elevated serum cytokines and immunoglobulins. In the absence of Zfp36l1 and Zfp36l2, the pool of cycling CTLA-4 in naïve Treg cells is reduced, Treg cells are less sensitive to IL-2 and IL-7 but are more sensitive to IFNγ. In mice lacking both RBP in Treg cells, the deletion of a single allele of Ifng is sufficient to ameliorate the pathology. Our results indicate that ZFP36L1 and ZFP36L2 regulate the availability of IFNγ and are required for the maintenance of Treg cell stability. Thus, ZFP36L1 and ZFP36L2 regulate multiple pathways that enable Treg cells to enforce immune homeostasis.
© 2025. The Author(s).
-
Genetics
-
Immunology and Microbiology
The tumor suppressor SALL2 opposes chemotherapeutic resistance in breast cancer.
In Molecular and Cellular Biochemistry on 1 May 2025 by Li, Q., Li, C., et al.
Chemotherapy continues to be the primary treatment for certain types of breast cancer. However, despite an initial positive response to chemotherapeutic agents, the development of resistance is inevitable. The exact molecular mechanisms underlying this phenomenon remain unclear. In this research, a significant downregulation of SALL2 expression was observed in chemo-resistant breast cancer, which was attributed to promoter methylation. Decreased SALL2 expression correlated significantly with poorer relapse-free survival in chemotherapy-treated patients with breast cancer. Functionally, SALL2 silencing induced a stem cell-like phenotype in breast cancer cells, fostering resistance to cisplatin both in vitro and in vivo. This resistance was mediated, at least in part, through the transcriptional regulation of BTG2, a negative regulator of stemness, achieved by direct binding to its promoter regions. These findings underscore the critical role of SALL2 in modulating cisplatin response and propose SALL2 as a potential prognostic biomarker for chemotherapy response in breast cancer.
© 2024. The Author(s).
-
Biochemistry and Molecular biology
-
Cancer Research
In Cancers on 26 March 2025 by Castaneda, M., den Hollander, P., et al.
Background: Aggressive forms of breast cancer, such as triple-negative breast cancer (TNBC), are associated with an increase in cancer cells that exhibit stem cell properties. The activation of the epithelial-mesenchymal transition (EMT) program, mediated by the transcription factor FOXC2, generates these stem-like cells. FOXC2 is linked to poor prognoses across various cancer types and is notably upregulated in TNBC, where it establishes and sustains these stem-like cells within the tumor population. Methods: Here, we decode the pathways regulating FOXC2 activation using EMT-enriched cell line models. Stemness was assessed using mammosphere assays and mesenchymal markers by western blot. Expression correlations with clinical data was examined using the EMTome. Results: We demonstrate that β-catenin serves as a critical mediator of mesenchymal and stemness characteristics through FOXC2 upregulation. By disrupting β-catenin, we find that FOXC2 expression, mesenchymal properties, and stemness are reduced; however, the introduction of exogenous FOXC2 expression in β-catenin deficient cells is enough to restore the mesenchymal and stemness phenotype. These findings support the idea that FOXC2 acts as the downstream regulator of β-catenin and influences both mesenchymal and stemness properties. Moreover, there is a positive correlation between the expression of β-catenin and FOXC2 in various cancer subtypes observed in clinical patient samples. Conclusions: Our study clarifies the role of the β-catenin/FOXC2 signaling axis in maintaining stemness properties, suggesting potential targets for TNBC and other cancers driven by EMT-related mesenchymal and stemness characteristics.
-
Stem Cells and Developmental Biology
Identification of a SNAI1 enhancer RNA that drives cancer cell plasticity.
In Nature Communications on 25 March 2025 by Fan, C., Wang, Q., et al.
Enhancer RNAs (eRNAs) are a pivotal class of enhancer-derived non-coding RNAs that drive gene expression. Here we identify the SNAI1 enhancer RNA (SNAI1e; SCREEM2) as a key activator of SNAI1 expression and a potent enforcer of transforming growth factor-β (TGF-β)/SMAD signaling in cancer cells. SNAI1e depletion impairs TGF-β-induced epithelial-mesenchymal transition (EMT), migration, in vivo extravasation, stemness, and chemotherapy resistance in breast cancer cells. SNAI1e functions as an eRNA to cis-regulate SNAI1 enhancer activity by binding to and strengthening the enrichment of the transcriptional co-activator bromodomain containing protein 4 (BRD4) at the local enhancer. SNAI1e selectively promotes the expression of SNAI1, which encodes the EMT transcription factor SNAI1. Furthermore, we reveal that SNAI1 interacts with and anchors the inhibitory SMAD7 in the nucleus, and thereby prevents TGF-β type I receptor (TβRI) polyubiquitination and proteasomal degradation. Our findings establish SNAI1e as a critical driver of SNAI1 expression and TGF-β-induced cell plasticity.
© 2025. The Author(s).
-
FC/FACS
-
Homo sapiens (Human)
-
Cancer Research
-
Genetics
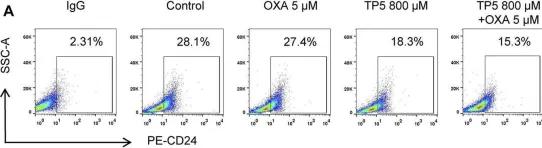
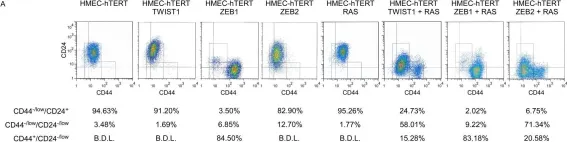
In Front Pharmacol on 5 March 2022 by Yu, P. C., Liu, D., et al.
Fig.7.A

-
FC/FACS
-
Collected and cropped from Front Pharmacol by CiteAb, provided under a CC-BY license
Image 1 of 5
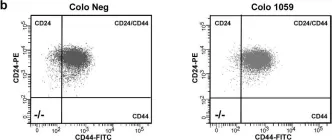
In Sci Rep on 17 May 2019 by Hocevar, B. A.
Fig.3.B

-
FC/FACS
-
Homo sapiens (Human)
Collected and cropped from Sci Rep by CiteAb, provided under a CC-BY license
Image 1 of 5
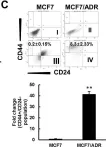
In Cancers (Basel) on 8 April 2019 by Lee, N. H., Park, S. R., et al.
Fig.1.D

-
FC/FACS
-
Collected and cropped from Cancers (Basel) by CiteAb, provided under a CC-BY license
Image 1 of 5
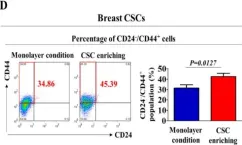
In Oncotarget on 19 September 2017 by Manandhar, S., Kim, C. G., et al.
Fig.1.C

-
FC/FACS
-
Homo sapiens (Human)
Collected and cropped from Oncotarget by CiteAb, provided under a CC-BY license
Image 1 of 5
In PLoS Genet on 2 June 2012 by Morel, A. P., Hinkal, G. W., et al.
Fig.5.A

-
FC/FACS
-
Collected and cropped from PLoS Genet by CiteAb, provided under a CC-BY license
Image 1 of 5