Terminal erythropoiesis is a complex multistep process involving coordination of gene transcription and dramatic nuclear condensation, which leads to the expulsion of nuclei to generate reticulocytes. However, we lack a comprehensive understanding of the key transcriptional and epigenetic regulators involved.
We used a high-throughput small molecule screen in primary CD34+-derived human erythroblasts to identify targets that promoted terminal erythropoiesis, and further confirmed the phenotype in different differentiation systems by inhibitors and shRNAs of different BRD4 isoforms. Then we performed RNA-seq, ATAC-seq, ChIP-qPCR, Co-IP, and reanalyzed previously-published transcriptional data and mass spectrometric data to clarify how BRD4 regulates terminal erythropoiesis.
We identified that inhibitors of the bromodomain protein BRD4, an epigenetic reader and transcriptional activator together with CDK9, promoted terminal erythropoiesis from hematopoietic stem/progenitor cells and embryonic stem cells, and enhanced enucleation. Combined analysis of our RNA-seq, ATAC-seq, and previously-published transcriptional data of erythroblast differentiation at different stages confirmed that BRD4 inhibition accelerates erythroblast maturation. Unexpectedly, this BRD4 function was independent of its classical CDK9 interaction and transcriptional activation. Instead, RNA-seq, ATAC-seq, and Cut&Tag upon BRD4 inhibition revealed that BRD4 regulates erythropoiesis by inhibiting the small G protein RhoB and disrupts actin reorganization. ChIP-qPCR, Co-IP, and functional studies revealed that BRD4 acts as a transcriptional repressor by interacting with the histone methyltransferase EHMT1/2.
We demonstrate a non-classical role for BRD4 as a transcriptional repressor of RhoB to regulate erythroid maturation, and classical CDK9 dependent role to regulate cell proliferation of erythroblasts. Besides, we clarify RhoB's activity and function during terminal erythropoiesis. BRD4 inhibition might be a simple method to promote in vitro blood cell production, and a candidate therapeutic target for diseases leading to dyserythropoiesis such as myelodysplastic syndromes.
© 2025. The Author(s).
Product Citations: 57
BRD4 acts as a transcriptional repressor of RhoB to inhibit terminal erythropoiesis.
In Journal of Hematology & Oncology on 1 July 2025 by Chen, Y., Huo, D., et al.
-
Biochemistry and Molecular biology
In International Journal of Stem Cells on 9 June 2025 by Park, J., Koh, H., et al.
Human pluripotent stem cells (hPSCs) can be used to investigate hematopoietic development and have the potential to advance cell-based therapies and to facilitate developmental biology studies. However, efficient ex vivo differentiation into hematopoietic lineages, including red blood cells (RBCs) of the erythroid lineage and immune cells such as macrophages of the myeloid lineage, is hampered by the need for precise temporal regulation of cytokines and growth factors. In this study, we developed an optimized protocol for hematopoietic lineage specification from hPSCs by fine-tuning the temporal dynamics of cytokine and growth factor applications. Prolonged mesodermal specification in the absence of hemogenic cytokines significantly enhanced the generation of hematopoietic progenitors (CD34+CD45+) with robust functional potential. Early administration of interleukin (IL)-3 during hematopoietic specification promoted progenitor expansion and maturation. Supplementation of bone morphogenetic protein 4 at the hematopoietic maturation stage enhanced the differentiation efficiency and preferentially drove myeloid lineage commitment toward macrophages at the expense of erythroid differentiation. The timing of erythropoietin administration was important in erythroid lineage commitment, and delayed treatment (day 10) enhanced erythroblast expansion and RBC production. By contrast, the timing of IL-6, GM-CSF, and M-CSF exposure did not significantly affect macrophage differentiation efficiency, suggesting that myeloid lineage specification follows a default pathway under optimized differentiation conditions. These findings suggest a refined, time-controlled strategy for directing hematopoietic differentiation from hPSCs, and provide insight into therapeutic blood cell production, regenerative medicine, and ex vivo modeling of hematopoietic disorders.
-
FC/FACS
-
Stem Cells and Developmental Biology
In Nature Communications on 4 September 2024 by Sun, S., Motazedian, A., et al.
Arterial endothelial cells (AECs) are the founder cells for intraembryonic haematopoiesis. Here, we report a method for the efficient generation of human haemogenic DLL4+ AECs from pluripotent stem cells (PSC). Time-series single-cell RNA-sequencing reveals the dynamic evolution of haematopoiesis and lymphopoiesis, generating cell types with counterparts present in early human embryos, including stages marked by the pre-haematopoietic stem cell genes MECOM/EVI1, MLLT3 and SPINK2. DLL4+ AECs robustly support lymphoid differentiation, without the requirement for exogenous NOTCH ligands. Using this system, we find IL7 acts as a morphogenic factor determining the fate choice between the T and innate lymphoid lineages and also plays a role in regulating the relative expression level of RAG1. Moreover, we document a developmental pathway by which human RAG1+ lymphoid precursors give rise to the natural killer cell lineage. Our study describes an efficient method for producing lymphoid progenitors, providing insights into their endothelial and haematopoietic ontogeny, and establishing a platform to investigate the development of the human blood system.
© 2024. The Author(s).
-
FC/FACS
Preprint on BioRxiv : the Preprint Server for Biology on 14 January 2024 by Frati, G., Brusson, M., et al.
Reactivation of fetal hemoglobin (HbF) expression through clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9-mediated disruption of regulatory elements involved in γ-globin gene repression is a promising gene therapy strategy for the treatment of sickle cell disease (SCD). However, preclinical studies aimed at optimizing the genome editing process and evaluating the safety of the editing strategy are necessary to translate this approach to the clinics. This is particularly relevant in the context of SCD, a disease characterized by inflammation, which can affect hematopoietic stem and progenitor cells (HSPCs), the target cell population in gene therapy approaches for hematopoietic disorders. Here, we describe a genome editing strategy leading to therapeutically relevant reactivation of HbF expression by targeting the binding sites (BSs) for the leukemia/lymphoma related factor (LRF) transcriptional repressor in the HBG1 and HBG2 γ-globin promoters. Electroporation of Cas9 ribonucleoprotein and single guide RNA (sgRNA) targeting the HBG promoters in healthy donor (HD) and patient-derived HSPCs resulted in a high frequency of LRF BS disruption and potent HbF synthesis in their erythroid progeny differentiated in vitro and ex vivo after transplantation into immunodeficient mice. LRF BS disruption did not impair SCD and HD HSPC engraftment and differentiation, but was more efficient in SCD than in HD cells. However, SCD HSPCs showed a reduced engraftment and a myeloid bias compared to HD cells. Importantly, in primary HSPCs, we detected off-target activity and the intra- and inter-chromosomal rearrangements between on- and off-target sites, which were more pronounced in SCD samples (likely because of the higher overall editing efficiency), but did not impact the target gene expression. Off-target activity was observed in vitro and in vivo, thus indicating that it does not impair engraftment and differentiation of both SCD and HD HSPCs. Finally, transcriptomic analyses showed that the genome editing procedure results in the upregulation of genes involved in DNA damage and inflammatory responses in both HD and SCD samples, although gene dysregulation was more evident in SCD HSPCs. Overall, this study provides evidences of feasibility, efficacy and safety for a genome editing strategy based on HbF reactivation and highlights the need of performing safety studies, when possible, in clinically relevant conditions, i.e., in patient-derived HSPCs.
In Genomics, Proteomics Bioinformatics on 1 December 2023 by Han, Y., Wang, S., et al.
The fetal liver (FL) is the key erythropoietic organ during fetal development, but knowledge on human FL erythropoiesis is very limited. In this study, we sorted primary erythroblasts from FL cells and performed RNA sequencing (RNA-seq) analyses. We found that temporal gene expression patterns reflected changes in function during primary human FL terminal erythropoiesis. Notably, the expression of genes enriched in proteolysis and autophagy was up-regulated in orthochromatic erythroblasts (OrthoEs), suggesting the involvement of these pathways in enucleation. We also performed RNA-seq of in vitro cultured erythroblasts derived from FL CD34+ cells. Comparison of transcriptomes between the primary and cultured erythroblasts revealed significant differences, indicating impacts of the culture system on gene expression. Notably, the expression of lipid metabolism-related genes was increased in cultured erythroblasts. We further immortalized erythroid cell lines from FL and cord blood (CB) CD34+ cells (FL-iEry and CB-iEry, respectively). FL-iEry and CB-iEry were immortalized at the proerythroblast stage and can be induced to differentiate into OrthoEs, but their enucleation ability was very low. Comparison of the transcriptomes between OrthoEs with and without enucleation capability revealed the down-regulation of pathways involved in chromatin organization and mitophagy in OrthoEs without enucleation capacity, indicating that defects in chromatin organization and mitophagy contribute to the inability of OrthoEs to enucleate. Additionally, the expression of HBE1, HBZ, and HBG2 was up-regulated in FL-iEry compared with CB-iEry, and such up-regulation was accompanied by down-regulated expression of BCL11A and up-regulated expression of LIN28B and IGF2BP1. Our study provides new insights into human FL erythropoiesis and rich resources for future studies.
Copyright © 2023 Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China. Published by Elsevier B.V. All rights reserved.
-
Homo sapiens (Human)
In Elife on 2 December 2022 by Sherpa, D., Mueller, J., et al.
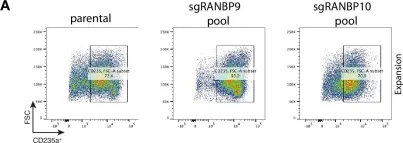
Fig.5.A

-
FC/FACS
-
Homo sapiens (Human)
Collected and cropped from Elife by CiteAb, provided under a CC-BY license
Image 1 of 4
In Elife on 2 December 2022 by Sherpa, D., Mueller, J., et al.
Fig.5.B

-
FC/FACS
-
Homo sapiens (Human)
Collected and cropped from Elife by CiteAb, provided under a CC-BY license
Image 1 of 4
In Elife on 2 December 2022 by Sherpa, D., Mueller, J., et al.
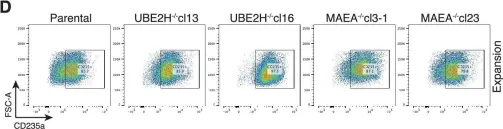
Fig.4.A

-
FC/FACS
-
Homo sapiens (Human)
Collected and cropped from Elife by CiteAb, provided under a CC-BY license
Image 1 of 4
In Elife on 2 December 2022 by Sherpa, D., Mueller, J., et al.
Fig.4.D

-
FC/FACS
-
Homo sapiens (Human)
Collected and cropped from Elife by CiteAb, provided under a CC-BY license
Image 1 of 4