Limited information exists regarding the impact of interferons (IFNs) on the information carried by extracellular vesicles (EVs). This study aimed at investigating whether IFN-α2b, IFN-β, IFN-γ, and IFN-λ1/2 modulate the content of EVs released by primary monocyte-derived macrophages (MDM). Small-EVs (sEVs) were purified by size exclusion chromatography from supernatants of MDM treated with IFNs. To characterize the concentration and dimensions of vesicles, nanoparticle tracking analysis was used. SEVs surface markers were examined by flow cytometry. IFN treatments induced a significant down-regulation of the exosomal markers CD9, CD63, and CD81 on sEVs, and a significant modulation of some adhesion molecules, major histocompatibility complexes and pro-coagulant proteins, suggesting IFNs influence biogenesis and shape the immunological asset of sEVs. SEVs released by IFN-stimulated MDM also impact lymphocyte function, showing significant modulation of lymphocyte activation and IL-17 release. Altogether, our results show that sEVs composition and activity are affected by IFN treatment of MDM.
© 2024 The Authors.
Product Citations: 38
In IScience on 21 June 2024 by Giannessi, F., Percario, Z., et al.
-
Homo sapiens (Human)
In Advanced Science (Weinheim, Baden-Wurttemberg, Germany) on 1 March 2024 by Li, H., Chiang, C. L., et al.
Detecting pancreatic duct adenocarcinoma (PDAC) in its early stages and predicting late-stage patient prognosis undergoing chemotherapy is challenging. This work shows that the activation of specific oncogenes leads to elevated expression of mRNAs and their corresponding proteins in extracellular vesicles (EVs) circulating in blood. Utilizing an immune lipoplex nanoparticle (ILN) biochip assay, these findings demonstrate that glypican 1 (GPC1) mRNA expression in the exosomes-rich (Exo) EV subpopulation and GPC1 membrane protein (mProtein) expression in the microvesicles-rich (MV) EV subpopulation, particularly the tumor associated microvesicles (tMV), served as a viable biomarker for PDAC. A combined analysis effectively discriminated early-stage PDAC patients from benign pancreatic diseases and healthy donors in sizable clinical from multiple hospitals. Furthermore, among late-stage PDAC patients undergoing chemotherapy, lower GPC1 tMV-mProtein and Exo-mRNA expression before treatment correlated significantly with prolonged overall survival. These findings underscore the potential of vesicular GPC1 expression for early PDAC screenings and chemotherapy prognosis.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
-
Cancer Research
-
Genetics
In EBioMedicine on 1 December 2023 by Han, B., Lv, Y., et al.
SARS-CoV-2 infects host cells via an ACE2/TMPRSS2 entry mechanism. Monocytes and macrophages, which play a key role during severe COVID-19 express only low or no ACE2, suggesting alternative entry mechanisms in these cells. In silico analyses predicted GRP78, which is constitutively expressed on monocytes and macrophages, to be a potential candidate receptor for SARS-CoV-2 virus entry.
Hospitalized COVID-19 patients were characterized regarding their pro-inflammatory state and cell surface GRP78 (csGRP78) expression in comparison to healthy controls. RNA from CD14+ monocytes of patients and controls were subjected to transcriptome analysis that was specifically complemented by bioinformatic re-analyses of bronchoalveolar lavage fluid (BALF) datasets of COVID-19 patients with a focus on monocyte/macrophage subsets, SARS-CoV-2 infection state as well as GRP78 gene expression. Monocyte and macrophage immunohistocytochemistry on GRP78 was conducted in post-mortem lung tissues. SARS-CoV-2 spike and GRP78 protein interaction was analyzed by surface plasmon resonance, GST Pull-down and Co-Immunoprecipitation. SARS-CoV-2 pseudovirus or single spike protein uptake was quantified in csGRP78high THP-1 cells.
Cytokine patterns, monocyte activation markers and transcriptomic changes indicated typical COVID-19 associated inflammation accompanied by upregulated csGRP78 expression on peripheral blood and lung monocytes/macrophages. Subsequent cell culture experiments confirmed an association between elevated pro-inflammatory cytokine levels and upregulation of csGRP78. Interaction of csGRP78 and SARS-CoV-2 spike protein with a dissociation constant of KD = 55.2 nM was validated in vitro. Infection rate analyses in ACE2low and GRP78high THP-1 cells showed increased uptake of pseudovirus expressing SARS-CoV-2 spike protein.
Our results demonstrate that csGRP78 acts as a receptor for SARS-CoV-2 spike protein to mediate ACE2-independent virus entry into monocytes.
Funded by the Sino-German-Center for Science Promotion (C-0040) and the Germany Ministry BMWi/K [DLR-grant 50WB1931 and RP1920 to AC, DM, TW].
Copyright © 2023 The Authors. Published by Elsevier B.V. All rights reserved.
-
FC/FACS
-
Biochemistry and Molecular biology
-
COVID-19
-
Immunology and Microbiology
In Cancer Immunology Research on 1 November 2023 by Zhang, C., Kadu, S., et al.
Natural killer (NK) cells are frequently expanded for the clinic using irradiated, engineered K562 feeder cells expressing a core transgene set of membrane-bound (mb) IL15 and/or mbIL21 together with 41BBL. Prior comparisons of mbIL15 to mbIL21 for NK expansion lack comparisons of key attributes of the resulting NK cells, including their high-dimensional phenotype, polyfunctionality, the breadth and potency of cytotoxicity, cellular metabolism, and activity in xenograft tumor models. Moreover, despite multiple rounds of K562 stimulation, studies of sequential use of mbIL15- and mbIL21-based feeder cells are absent. We addressed these gaps and found that using mbIL15- versus mbIL21-based feeder cells drove distinct phenotypic and functional profiles. Feeder cells expressing mbIL15 alone drove superior functionality by nearly all measures, whereas those expressing mbIL21 alone drove superior yield. In combination, most attributes resembled those imparted by mbIL21, whereas in sequence, NK yield approximated that imparted by the first cytokine, and the phenotype, transcriptome, and function resembled that driven by the second cytokine, highlighting the plasticity of NK cell differentiation. The sequence mbIL21 followed by mbIL15 was advantageous in achieving significant yields of highly functional NK cells that demonstrated equivalent in vivo activity to those expanded by mbIL15 alone in two of three xenograft models. Our findings define the impact of mbIL15 versus mbIL21 during NK expansion and reveal a previously underappreciated tradeoff between NK yield and function for which sequential use of mbIL21-based followed by mbIL15-based feeder cells may be the optimal approach in many settings.
©2023 The Authors; Published by the American Association for Cancer Research.
-
FC/FACS
-
Homo sapiens (Human)
In Cell Rep Methods on 22 May 2023 by Mattsson, J., Ljungars, A., et al.
Phenotypic drug discovery (PDD) enables the target-agnostic generation of therapeutic drugs with novel mechanisms of action. However, realizing its full potential for biologics discovery requires new technologies to produce antibodies to all, a priori unknown, disease-associated biomolecules. We present a methodology that helps achieve this by integrating computational modeling, differential antibody display selection, and massive parallel sequencing. The method uses the law of mass action-based computational modeling to optimize antibody display selection and, by matching computationally modeled and experimentally selected sequence enrichment profiles, predict which antibody sequences encode specificity for disease-associated biomolecules. Applied to a phage display antibody library and cell-based antibody selection, ∼105 antibody sequences encoding specificity for tumor cell surface receptors expressed at 103-106 receptors/cell were discovered. We anticipate that this approach will be broadly applicable to molecular libraries coupling genotype to phenotype and to the screening of complex antigen populations for identification of antibodies to unknown disease-associated targets.
© 2023 The Author(s).
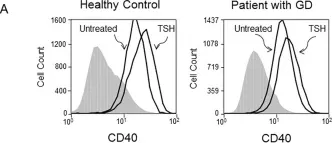
In PLoS One on 16 September 2016 by Mester, T., Raychaudhuri, N., et al.
Fig.1.A

-
FC/FACS
-
Homo sapiens (Human)
Collected and cropped from PLoS One by CiteAb, provided under a CC-BY license
Image 1 of 1