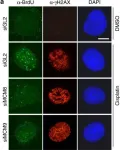
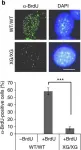
Zika virus (ZIKV) infection in human neural progenitors triggers DNA damage and activates DNA damage response, leading to cell cycle arrest that can retard brain development. Here, we link the ZIKV-induced S phase arrest to replication fork stalling and R-loop induction. DRIP-seq reveals that ZIKV infection induces R loops at specific loci strongly enriched in the interferon (IFN)-stimulated genes (ISGs). Bromouridine sequencing results further indicate that nascent ISGs transcripts are prone to R-loop induction upon infection. Knockout of IFN receptor eliminated the R loops on ISGs and partially rescued S-phase arrest in infected cells. And overexpression of RNaseH1 reduced ZIKV-mediated DNA damage and cell cycle arrest. We conclude that unscheduled expression of ISGs induced by ZIKV alters R-loop homeostasis and perturbs replication fork progression, leading to fork stalling and eventually DNA damage. IFN-dependent R-loop induction represents a previously unknown, nucleic acid-based mechanism for cell cycle arrest.
© The Author(s) 2025. Published by Oxford University Press on behalf of National Academy of Sciences.
Product Citations: 367
Interferon-dependent R-loop induction by Zika virus contributes to growth attenuation.
In PNAS Nexus on 1 May 2025 by Zhao, Y., Metzler, A. D., et al.
-
Immunology and Microbiology
Preprint on BioRxiv : the Preprint Server for Biology on 24 April 2025 by Urban, J. A., Ringwalt, D., et al.
ABSTRACT Stem cells have the unique ability to self-renew and differentiate into specialized cell types. Epigenetic mechanisms, including histones and their post-translational modifications, play a crucial role in regulating programs integral to a cell’s identity, like gene expression and DNA replication. However, the transcriptional, chromatin, and replication timing profiles of adult stem cells in vivo remain poorly understood. Containing germline stem cells (GSCs) and somatic cyst stem cells (CySCs), the Drosophila testis provides an excellent in vivo model for studying adult stem cells. However, the small number of stem cells and cellular heterogeneity of this tissue have limited comprehensive genomic studies. In this study, we developed cell type-specific genomic techniques to analyze the transcriptome, histone modification patterns, and replication timing of GSC-like and CySC-like cells. Single cell RNA-sequencing validated previous findings on GSC-CySC intercellular communication and revealed high expression of chromatin regulators in GSCs. To characterize chromatin landscapes, we developed a cell-type-specific chromatin profiling assay to map H3K4me3-, H3K27me3-, and H3K9me3-enriched regions, corresponding to euchromatic, facultative heterochromatic, and constitutive heterochromatic domains, respectively. Finally, we determined cell type-specific replication timing profiles, integrating our in vivo datasets with published data using cultured cell lines. Our results reveal that GSCs display a distinct replication program compared to somatic lineages, that aligns with chromatin state differences. Collectively, our integrated transcriptomic, chromatin, and replication datasets provide a comprehensive framework for understanding genome regulation differences between these in vivo stem cell populations, demonstrating the power of multi-omics in uncovering cell type-specific regulatory features.
-
Genetics
-
Stem Cells and Developmental Biology
In Science Advances on 11 April 2025 by Yang, Y., Wang, T. Y., et al.
Mounting evidence indicates that long noncoding RNAs (lncRNAs) play vital roles in tumorigenesis and progression of cancers. However, the functions and regulatory mechanisms of lncRNAs in prostate cancer (PCa) are still largely unknown. In this study, we found an lncRNA, PCa-associated transcript 71 (PRCAT71), highly expressed in metastatic and primary PCa compared to benign prostate tissues. Silencing PRCAT71 inhibited cancerous properties of PCa cells and androgen receptor (AR) signaling. Mechanistically, PRCAT71 acts as a scaffold to recruit K homology (KH)-type splicing regulatory protein (KHSRP) to AR messenger RNA (mRNA) and stabilize AR mRNA, leading to activated AR signaling. KHSRP plays a critical role in PCa progression. PRCAT71 is transcriptionally regulated by AR-driven enhancers, forming a positive regulatory loop between AR and PRCAT71 in PCa. Our study demonstrates a coordinated regulation of AR mRNA by lncRNA PRCAT71 and RNA binding protein KHSRP and provides insight that the PRCAT71-KHSRP-AR axis is a promising therapeutic target for treating PCa.
-
Cancer Research
-
Endocrinology and Physiology
Long-range enhancer-controlled genes are hypersensitive to regulatory factor perturbations.
In Cell Genom on 12 March 2025 by Tjalsma, S. J. D., Rinzema, N. J., et al.
Cell-type-specific gene activation is regulated by enhancers, sometimes located at large genomic distances from target gene promoters. Whether distal enhancers require specific factors to orchestrate gene regulation remains unclear. Here, we used enhancer distance-controlled reporter screens to find candidate factors. We depleted them and employed activity-by-contact predictions to genome-wide classify genes based on enhancer distance. Predicted distal enhancers typically control tissue-restricted genes and often are strong enhancers. We find cohesin, but also mediator, most specifically required for long-range activation, with cohesin repressing short-range gene activation and prioritizing distal over proximal HBB genes competing for shared enhancers. Long-range controlled genes are also most sensitive to perturbations of other regulatory proteins and to BET inhibitor JQ1, this being more a consequence of their distinct enhancer features than distance. Our work predicts that lengthening of intervening sequences can help limit the expression of target genes to specialized cells with optimal trans-factor environments.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Preprint on BioRxiv : the Preprint Server for Biology on 11 March 2025 by Trinh, A., Akhtar, N., et al.
Faithful epigenetic inheritance across cell divisions is essential to maintaining cell identity and involves numerous epigenetic modifications, whose roles in establishing chromatin architecture are less understood. Technological approaches to temporally order epigenetic modifications throughout the cell cycle often face limitations in sequence resolution and rely on potentially damaging mitotic labeling or conversion steps. Herein, we present M ethylation P seudotime A nalysis T hrough read-level H eterogeneity (MPATH), a label- and conversion-free method to infer post-replication DNA strand maturity from methylation patterns across single molecules. We use MPATH to temporally order hydroxymethylation throughout mitotic inheritance, revealing that CpGs within cis-regulatory elements undergo transitions between methylation states at sub-cell-cycle timescales. When applied to long reads generated by NOMe-seq, MPATH uncovered relationships between nucleosome occupancy and DNA maturity. Finally, extension of MPATH to phased reads reveals allele-specific trends in pseudotime distribution associated with X chromosome activity. Our findings suggest that when coupled with multimodal sequencing strategies, MPATH could provide valuable insights into chromatin restoration dynamics.
In Nat Commun on 7 June 2019 by Delaney, K., Strobino, M., et al.
Fig.1.E

-
ICC-IF
-
Caenorhabditis elegans
Collected and cropped from Nat Commun by CiteAb, provided under a CC-BY license
Image 1 of 6
In Development on 27 November 2018 by Samarajeewa, A., Lenz, D. R., et al.
Fig.2.A

-
IF
-
Mus musculus (House mouse)
Collected and cropped from Development by CiteAb, provided under a CC-BY license
Image 1 of 6
In Development on 27 November 2018 by Samarajeewa, A., Lenz, D. R., et al.
Fig.1.A

-
IF
-
Mus musculus (House mouse)
Collected and cropped from Development by CiteAb, provided under a CC-BY license
Image 1 of 6
In Development on 27 November 2018 by Samarajeewa, A., Lenz, D. R., et al.
Fig.5.A

-
IF
-
Mus musculus (House mouse)
Collected and cropped from Development by CiteAb, provided under a CC-BY license
Image 1 of 6
In Nat Commun on 28 July 2015 by Lee, K. Y., Im, J. S., et al.
Fig.2.A

-
ICC-IF
-
Mus musculus (House mouse)
Collected and cropped from Nat Commun by CiteAb, provided under a CC-BY license
Image 1 of 6
In Nat Commun on 28 July 2015 by Lee, K. Y., Im, J. S., et al.
Fig.2.B

-
ICC-IF
-
Mus musculus (House mouse)
Collected and cropped from Nat Commun by CiteAb, provided under a CC-BY license
Image 1 of 6