The tumor suppressor TP53 is frequently inactivated in a mutation-independent manner in cancers and is reactivated by inhibiting its negative regulators. We here cotarget MDM2 and the nuclear exporter XPO1 to maximize transcriptional activity of p53. MDM2/XPO1 inhibition accumulated nuclear p53 and elicited a 25- to 60-fold increase of its transcriptional targets. TP53 regulates MYC, and MDM2/XPO1 inhibition disrupted the c-MYC-regulated transcriptome, resulting in the synergistic induction of apoptosis in acute myeloid leukemia (AML). Unexpectedly, venetoclax-resistant AMLs express high levels of c-MYC and are vulnerable to MDM2/XPO1 inhibition in vivo. However, AML cells persisting after MDM2/XPO1 inhibition exhibit a quiescence- and stress response-associated phenotype. Venetoclax overcomes that resistance, as shown by single-cell mass cytometry. The triple inhibition of MDM2, XPO1, and BCL2 was highly effective against venetoclax-resistant AML in vivo. Our results propose a novel, highly translatable therapeutic approach leveraging p53 reactivation to overcome nongenetic, stress-adapted venetoclax resistance.
Product Citations: 47
In Science Advances on 1 December 2023 by Nishida, Y., Ishizawa, J., et al.
-
Biochemistry and Molecular biology
Preprint on BioRxiv : the Preprint Server for Biology on 17 June 2022 by Silberberg, G., Walling, B., et al.
Despite considerable progress made in improving therapeutic strategies, the overall survival for patients diagnosed with various cancer types remains low. Further, patients often cycle through multiple therapeutic options before finding an effective regimen for the specific malignancy being treated. A focus on building enhanced computational models, which prioritize therapeutic regimens based on a tumor’s complete molecular profile, will improve the patient experience and augment initial outcomes. In this study, we present an integrative analysis of multiple omic datasets coupled with phenotypic and therapeutic response profiles of Cytarabine from a cohort of primary AML tumors, and Olaparib from a cohort of Patient-Derived Xenograft (PDX) models of ovarian cancer. These analyses, termed P harmaco- P heno- M ulti o mic (PPMO) Integration, established novel complex biomarker profiles that were used to accurately predict prospective therapeutic response profiles in cohorts of newly profiled AML and ovarian tumors. Results from the computational analyses also provide new insights into disease etiology and the mechanisms of therapeutic resistance. Collectively, this study provides proof-of-concept in the use of PPMO to establish highly accurate predictive models of therapeutic response, and the power of leveraging this method to unveil cancer disease mechanisms.
-
Cancer Research
In Nature Communications on 19 May 2022 by Baran, N., Lodi, A., et al.
T-cell acute lymphoblastic leukemia (T-ALL) is commonly driven by activating mutations in NOTCH1 that facilitate glutamine oxidation. Here we identify oxidative phosphorylation (OxPhos) as a critical pathway for leukemia cell survival and demonstrate a direct relationship between NOTCH1, elevated OxPhos gene expression, and acquired chemoresistance in pre-leukemic and leukemic models. Disrupting OxPhos with IACS-010759, an inhibitor of mitochondrial complex I, causes potent growth inhibition through induction of metabolic shut-down and redox imbalance in NOTCH1-mutated and less so in NOTCH1-wt T-ALL cells. Mechanistically, inhibition of OxPhos induces a metabolic reprogramming into glutaminolysis. We show that pharmacological blockade of OxPhos combined with inducible knock-down of glutaminase, the key glutamine enzyme, confers synthetic lethality in mice harboring NOTCH1-mutated T-ALL. We leverage on this synthetic lethal interaction to demonstrate that IACS-010759 in combination with chemotherapy containing L-asparaginase, an enzyme that uncovers the glutamine dependency of leukemic cells, causes reduced glutaminolysis and profound tumor reduction in pre-clinical models of human T-ALL. In summary, this metabolic dependency of T-ALL on OxPhos provides a rational therapeutic target.
© 2022. The Author(s).
-
Biochemistry and Molecular biology
-
Cancer Research
-
Cell Biology
-
Immunology and Microbiology
In Nature Communications on 11 May 2022 by Stern, L., McGuire, H. M., et al.
Human cytomegalovirus reactivation is a major opportunistic infection after allogeneic haematopoietic stem cell transplantation and has a complex relationship with post-transplant immune reconstitution. Here, we use mass cytometry to define patterns of innate and adaptive immune cell reconstitution at key phases of human cytomegalovirus reactivation in the first 100 days post haematopoietic stem cell transplantation. Human cytomegalovirus reactivation is associated with the development of activated, memory T-cell profiles, with faster effector-memory CD4+ T-cell recovery in patients with low-level versus high-level human cytomegalovirus DNAemia. Mucosal-associated invariant T cell levels at the initial detection of human cytomegalovirus DNAemia are significantly lower in patients who subsequently develop high-level versus low-level human cytomegalovirus reactivation. Our data describe distinct immune signatures that emerged with human cytomegalovirus reactivation after haematopoietic stem cell transplantation, and highlight Mucosal-associated invariant T cell levels at the first detection of reactivation as a marker that may be useful to anticipate the magnitude of human cytomegalovirus DNAemia.
© 2022. The Author(s).
-
Immunology and Microbiology
-
Stem Cells and Developmental Biology
In European Journal of Immunology on 1 July 2021 by Campana, S., De Pasquale, C., et al.
Human CD117+ CRTH2neg innate lymphoid cells (ILC) comprise multipotent precursors (ILCp), which are able to differentiate into subtypes in response to different signals received in peripheral tissues. NKp46+ ILCp have been reported to associate with ILC3 whereas KLRG1+ ILCp with ILC2, although the latter can also generate other ILC subsets, thus, maintaining a substantial plasticity. We here showed that CD62L is expressed by ILCp exclusively within KLRG1+ population and its expression marks a loss of their broad differentiation potential. Analysis of cytokine production and relevant markers demonstrated that CD62L+ ILCp mainly differentiate into ILC2 whereas CD62Lneg counterpart can also differentiate into other ILC subsets depending on the signals they receive. Remarkably, in peripheral blood of psoriatic patients, where ILC3 are usually enriched, CD62L+ ILC were drastically reduced, whereas CD62Lneg ILC2 upregulated both RORγt and NKp46, thus, suggesting an ongoing conversion to ILC3. Therefore, CD62L now emerges as a potential marker to identify a skewing toward type 2 among ILCp.
© 2021 The Authors. European Journal of Immunology published by Wiley-VCH GmbH.
-
Immunology and Microbiology
In J Transl Med on 15 November 2016 by Seebach, C., Henrich, D., et al.
Fig.1.A
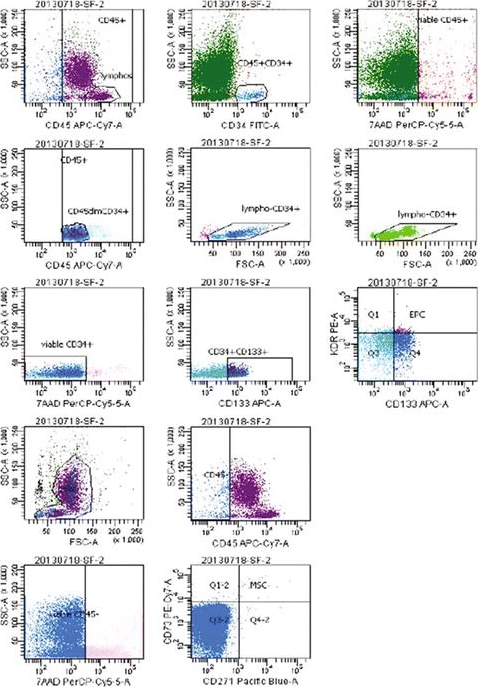
-
FC/FACS
-
Homo sapiens (Human)
Collected and cropped from Journal of Translational Medicine by CiteAb, provided under a CC-BY license
Image 1 of 1
