HIV-1 Nef enhances virus propagation by down-regulating CD4 and SERINC5. However, recent evidence points to the existence of an additional Nef-sensitive restriction mechanism. We now show that Nef suppresses the aberrant cleavage of HIV-1 gp41 by ADAM10, a virion-associated cellular ectodomain sheddase, and thus increases the amount of HIV-1 envelope glycoprotein (Env) on virions. Additionally, Nef inhibits the shedding of at least some cellular ADAM10 substrates, resulting in their accumulation on HIV-1 virions. Whereas Nef+ HIV-1 replicated only marginally better in the absence of ADAM10, the propagation of Nef- HIV-1 was notably rescued in ADAM10- T cell lines. Crucially, Nef- HIV-1 also benefited from the absence of ADAM10 in primary CD4+ T cells. Collectively, our results indicate that ADAM10 negatively affects both laboratory-adapted and primary HIV-1 strains by shedding the ectodomains of viral and cellular transmembrane proteins from virions and that Nef rescues virus replication by counteracting ADAM10.
Product Citations: 85
The ectodomain sheddase ADAM10 restricts HIV-1 propagation and is counteracted by Nef.
In Science Advances on 18 April 2025 by Olety, B., Usami, Y., et al.
Innate cell markers that predict anti-HIV neutralizing antibody titers in vaccinated macaques.
In Cell Reports Medicine on 18 October 2022 by Van Tilbeurgh, M., Maisonnasse, P., et al.
Given the time and resources invested in clinical trials, innovative prediction methods are needed to decrease late-stage failure in vaccine development. We identify combinations of early innate responses that predict neutralizing antibody (nAb) responses induced in HIV-Env SOSIP immunized cynomolgus macaques using various routes of vaccine injection and adjuvants. We analyze blood myeloid cells before and 24 h after each immunization by mass cytometry using a three-step clustering, and we discriminate unique vaccine signatures based on HLA-DR, CD39, CD86, CD11b, CD45, CD64, CD14, CD32, CD11c, CD123, CD4, CD16, and CADM1 surface expression. Various combinations of these markers characterize cell families positively associated with nAb production, whereas CADM1-expressing cells are negatively associated (p < 0.05). Our results demonstrate that monitoring immune signatures during early vaccine development could assist in identifying biomarkers that predict vaccine immunogenicity.Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
HIV-1 propagation is highly dependent on basal levels of the restriction factor BST2.
In Science Advances on 29 October 2021 by Olety, B., Peters, P., et al.
BST2 is an interferon-inducible antiviral host protein antagonized by HIV-1 Vpu that entraps nascent HIV-1 virions on the cell surface. Unexpectedly, we find that HIV-1 lacking Nef can revert to full replication competence simply by losing the ability to antagonize BST2. Using gene editing together with cell sorting, we demonstrate that even the propagation of wild-type HIV-1 is strikingly dependent on BST2, including in primary human cells. HIV-1 propagation in BST2−/− populations can be fully rescued by exogenous BST2 irrespective of its capacity to signal and even by an artificial BST2-like protein that shares its virion entrapment activity but lacks sequence homology. Counterintuitively, our results reveal that HIV-1 propagation is critically dependent on basal levels of virion tethering by a key component of innate antiviral immunity.
In Frontiers in Immunology on 6 July 2021 by Huot, N., Rascle, P., et al.
CD4 T cell responses constitute an important component of adaptive immunity and are critical regulators of anti-microbial protection. CD4+ T cells expressing CD32a have been identified as a target for HIV. CD32a is an Fcγ receptor known to be expressed on myeloid cells, granulocytes, B cells and NK cells. Little is known about the biology of CD32+CD4+ T cells. Our goal was to understand the dynamics of CD32+CD4+ T cells in tissues. We analyzed these cells in the blood, lymph nodes, spleen, ileum, jejunum and liver of two nonhuman primate models frequently used in biomedical research: African green monkeys (AGM) and macaques. We studied them in healthy animals and during viral (SIV) infection. We performed phenotypic and transcriptomic analysis at different stages of infection. In addition, we compared CD32+CD4+ T cells in tissues with well-controlled (spleen) and not efficiently controlled (jejunum) SIV replication in AGM. The CD32+CD4+ T cells more frequently expressed markers associated with T cell activation and HIV infection (CCR5, PD-1, CXCR5, CXCR3) and had higher levels of actively transcribed SIV RNA than CD32-CD4+T cells. Furthermore, CD32+CD4+ T cells from lymphoid tissues strongly expressed B-cell-related transcriptomic signatures, and displayed B cell markers at the cell surface, including immunoglobulins CD32+CD4+ T cells were rare in healthy animals and blood but increased strongly in tissues with ongoing viral replication. CD32+CD4+ T cell levels in tissues correlated with viremia. Our results suggest that the tissue environment induced by SIV replication drives the accumulation of these unusual cells with enhanced susceptibility to viral infection.
Copyright © 2021 Huot, Rascle, Planchais, Contreras, Passaes, Le Grand, Beignon, Kornobis, Legendre, Varet, Saez-Cirion, Mouquet, Jacquelin and Müller-Trutwin.
-
Immunology and Microbiology
Highly Efficient Generation of Transgenically Augmented CAR NK Cells Overexpressing CXCR4.
In Frontiers in Immunology on 29 September 2020 by Jamali, A., Hadjati, J., et al.
Natural killer (NK) cells are a noteworthy lymphocyte subset in cancer adoptive cell therapy. NK cells initiate innate immune responses against infections and malignancies with natural cytotoxicity, which is independent of foreign antigen recognition. Based on these substantive features, genetically modifying NK cells is among the prime goals in immunotherapy but is currently difficult to achieve. Recently, we reported a fully human CAR19 construct (huCAR19) with remarkable function in gene-modified T-cells. Here, we show efficient and stable gene delivery of huCAR19 to primary human NK cells using lentiviral vectors with transduction efficiencies comparable to those achieved with NK cell lines. These huCAR19 NK cells display specific and potent cytotoxic activity against target cells. To improve homing of NK cells to the bone marrow, we augmented huCAR19 NK cells with the human CXCR4 gene, resulting in transgenically augmented CAR NK cells (TRACKs). Compared to conventional CAR NK cells, TRACKs exhibit enhanced migration capacity in response to recombinant SDF-1 or bone marrow stromal cells while retaining functional and cytolytic activity against target cells. Based on these promising findings, TRACKs may become a novel candidate for immunotherapeutic strategies in clinical applications.
Copyright © 2020 Jamali, Hadjati, Madjd, Mirzaei, Thalheimer, Agarwal, Bonig, Ullrich and Hartmann.
-
Immunology and Microbiology
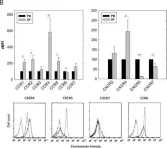
In Arthritis Res Ther on 7 June 2018 by Armas-González, E., Domínguez-Luis, M. J., et al.
Fig.1.B

-
FC/FACS
-
Collected and cropped from Arthritis Res Ther by CiteAb, provided under a CC-BY license
Image 1 of 2
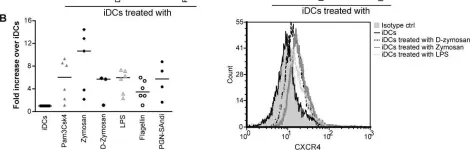
In PLoS One on 12 July 2013 by Côté, S. C., Plante, A., et al.
Fig.5.B

-
FC/FACS
-
Collected and cropped from PLoS One by CiteAb, provided under a CC-BY license
Image 1 of 2