Molecular subtyping of small-cell lung cancer (SCLC) has major implications for prognostic relevance and treatment guidance. This study aimed to explore the feasibility of a novel tracer targeting C-X-C-chemokine-receptor-type-4 (CXCR4) for distinguishing different SCLC subtypes.
Thirty-five patients with pathologically confirmed SCLC were enrolled in this prospective study. Immunohistochemical staining was performed to classify the molecular subtypes into SCLC-A, SCLC-N, SCLC-P, and SCLC-I. [¹⁸F]AlF-NOTA-QHY-04 PET/CT parameters were obtained, including the maximum, mean, and peak standard uptake values (SUVmax, SUVmean, and SUVpeak, respectively) and the ratios of tumors (T) and normal tissues (NT) based on the SUVmax (T/NT). These parameters were compared among the molecular subtypes. A receiver operating characteristic (ROC) curve was used to analyze the performance of the parameters for distinguishing SCLC-N from other subtypes and neuroendocrine (NE) subtypes (SCLC-A and SCLC-N) from non-NE subtypes (SCLC-P and SCLC-I).
The molecular subtypes were SCLC-A (n = 17), SCLC-N (n = 6), SCLC-P (n = 7), and SCLC-I (n = 5). The SCLC-N subtype exhibited significantly higher uptake in both primary tumors and lymph node metastases than the other three subtypes (P < 0.05). When SCLC-N was compared with the other three subtypes combined (referred to as "other SCLCs"), all parameters were significantly higher in the SCLC-N group (P < 0.05). ROC analysis showed that these parameters had high accuracy in distinguishing SCLC-N from other SCLCs (area under ROC curve: 0.868-0.948 for primary tumors and 0.783-0.888 for lymph node metastases). Compared with the non-NE group, the SUVmax, SUVmean, and T/NTlung were significantly higher in the NE group for primary tumors. ROC analysis showed moderate accuracy in distinguishing between the NE and non-NE groups (ROC area: 0.692-0.786 for primary tumors and 0.692-0.815 for lymph node metastases).
Our preliminary findings indicate that CXCR4-directed PET/CT imaging using [¹⁸F]AlF-NOTA-QHY-04 may differentiate between SCLC-N and other molecular subtypes and between NE and non-NE subtypes of SCLC.
Copyright © 2025 The Korean Society of Radiology.
Product Citations: 260
In Korean Journal of Radiology : Official Journal of the Korean Radiological Society on 1 June 2025 by Luo, Y., Cheng, K., et al.
-
Cancer Research
In Science Advances on 25 April 2025 by Brumage, L., Best, S., et al.
Exquisitely chemosensitive initially, small cell lung cancer (SCLC) exhibits dismal outcomes owing to rapid transition to chemoresistance. Elucidating the genetic underpinnings has been challenging owing to limitations with cellular models. As SCLC patient-derived xenograft (PDX) models mimic therapeutic responses, we perform genetic screens in chemosensitive PDX models to identify drivers of chemoresistance. cDNA overexpression screens identify MYC, MYCN, and MYCL, while CRISPR deletion screens identify KEAP1 loss as driving chemoresistance. Deletion of KEAP1 switched a chemosensitive SCLC PDX model to become chemoresistant and resulted in sensitivity to inhibition of glutamine metabolism. Data from the IMpower133 clinical trial revealed ~6% of patients with extensive-stage SCLC exhibit KEAP1 genetic alterations, with activation of a KEAP1/NRF2 transcriptional signature associated with reduced survival upon chemotherapy treatment. While roles for KEAP1/NRF2 have been unappreciated in SCLC, our genetic screens revealed KEAP1 loss as a driver of chemoresistance, while patient genomic analyses demonstrate clinical importance.
-
Cancer Research
In Cancer Metabolism on 31 March 2025 by Solta, A., Ernhofer, B., et al.
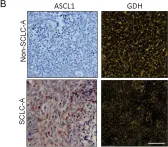
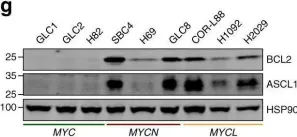
Small cell lung cancer (SCLC) is an aggressive malignancy with distinct molecular subtypes defined by transcription factors and inflammatory characteristics. This follow-up study aimed to validate the unique metabolic phenotype in achaete-scute homologue 1 (ASCL1)-driven SCLC cell lines and human tumor tissue.
Metabolic alterations were analyzed using proteomic data. Structural and functional differences of mitochondria were investigated using qPCR, flow cytometry, confocal imaging, and transmission electron microscopy and seahorse assays. Several metabolic inhibitors were tested using MTT-based and clonogenic assays. Single-cell enzyme activity assays were conducted on cell lines and tumor tissue samples of SCLC patients.
We found increased mitochondrial numbers correlating with higher oxidative phosphorylation activity in ASCL1-dominant cells compared to other SCLC subtypes. Metabolic inhibitors targeting mitochondrial respiratory complex-I or carnitine palmitoyltransferase 1 revealed higher responsiveness in SCLC-A. Conversely, we demonstrated that non-ASCL1-driven SCLCs with lower oxidative signatures show dependence on glutaminolysis as evidenced by the enhanced susceptibility to glutaminase inhibition. Accordingly, we detected increased glutamate-dehydrogenase activity in non-ASCL1-dominant cell lines as well as in human SCLC tissue samples.
Distinct SCLC subtypes exhibit unique metabolic vulnerabilities, suggesting potential for subtype-specific therapies targeting the respiratory chain, fatty acid transport, or glutaminolysis.
© 2025. The Author(s).
-
IHC
-
Cancer Research
-
Cell Biology
Outer radial glia promotes white matter regeneration after neonatal brain injury.
In Cell Reports Medicine on 18 March 2025 by Jinnou, H., Rosko, L. M., et al.
The developing gyrencephalic brain contains a large population of neural stem cells in the ventricular zone and outer subventricular zone (OSVZ), the latter populated by outer radial glia (oRG). The role of oRG during postnatal development is not well understood. We show that oRG cells increase proliferative capacity and contribute to oligodendrocyte precursor cell (OPC) production following brain injury in human infants and neonatal piglets, whose brains resemble the human brain in structure and development. RNA sequencing revealed oRG-specific transcriptional responses to injury in piglets and showed that the activating transcription factor 5 (ATF5) pathway positively regulates oRG proliferation. Intranasal activation of ATF5 using salubrinal enhanced OSVZ-derived oligodendrogenesis in the injured periventricular white matter and improved functional recovery. These results reveal a key role for postnatal oRG in brain injury recovery and identify ATF5 as a potential therapeutic target for treating white matter injury in infants.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
-
Neuroscience
In Molecular Biology Reports on 12 March 2025 by Hanspal, M. A., Presland, J., et al.
Achaete-Scute complex homolog 1 (ASCL1) is a multi-faceted pro-neural transcription factor, playing a role in several processes during embryonic development and into adulthood, including neural progenitor proliferation and neuronal differentiation. This versatility is achieved through tightly controlled expression of ASCL1, either via integrating intracellular signalling cues or stabilisation at the protein level. The role of kinases in ASCL1-mediated neurogenesis is emerging, but to date few kinases have been attributed to act directly or indirectly on ASCL1.
To address this, we designed a cell-based high-throughput screen to identify kinase inhibitors that enhance ASCL1 protein levels. From this screen, two kinase inhibitors were identified to increase ASCL1 stability and transcriptional activity, and subsequent validation indicated that the effect was driven indirectly through Janus kinase family members.
These compounds may serve as useful tools for further investigating the role played by kinases in regulating neurogenesis and ultimately enable better understanding of how ASCL1 integrates different signalling cues to orchestrate with high precision the differentiation of progenitor cells into neurons.
© 2025. Merck & Co., Inc., Rahway, NJ, USA and its affiliates.
-
WB
-
Biochemistry and Molecular biology
In Cancer Metab on 31 March 2025 by Solta, A., Ernhofer, B., et al.
Fig.2.B

-
IHC
-
Collected and cropped from Cancer Metab by CiteAb, provided under a CC-BY license
Image 1 of 7
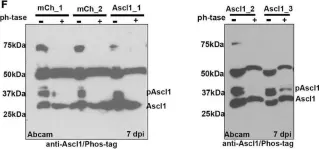
In Front Neurosci on 6 September 2022 by Ghazale, H., Park, E., et al.
Fig.1.F

-
WB
-
Collected and cropped from Front Neurosci by CiteAb, provided under a CC-BY license
Image 1 of 7
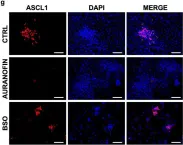
In Nat Commun on 6 April 2021 by Bebber, C. M., Thomas, E., et al.
Fig.5.G

-
ICC-IF
-
Collected and cropped from Nat Commun by CiteAb, provided under a CC-BY license
Image 1 of 7
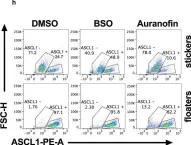
In Nat Commun on 6 April 2021 by Bebber, C. M., Thomas, E., et al.
Fig.5.H

-
FC/FACS
-
Collected and cropped from Nat Commun by CiteAb, provided under a CC-BY license
Image 1 of 7
In Nat Commun on 2 August 2019 by Dammert, M. A., Brägelmann, J., et al.
Fig.1.G

-
WB
-
Collected and cropped from Nat Commun by CiteAb, provided under a CC-BY license
Image 1 of 7
In EMBO J on 15 March 2019 by Bragado Alonso, S., Reinert, J. K., et al.
Fig.1.B

-
IHC-IF
-
Mus musculus (House mouse)
Collected and cropped from EMBO J by CiteAb, provided under a CC-BY license
Image 1 of 7
In Aging Dis on 1 May 2017 by Long, Q., Luo, Q., et al.
Fig.2.A

-
WB
-
Rattus norvegicus (Rat)
Collected and cropped from Aging Dis by CiteAb, provided under a CC-BY license
Image 1 of 7