Loss-of-function (LOF) variants in IL6ST, encoding GP130, can cause hyper-IgE syndrome (HIES). Monoallelic LOF variants in IL6ST lead to HIES when located in the intracellular domain downstream of box 1/2 and upstream of the STAT3 phosphorylation sites and the recycling motif, due to their dominant negative (DN) activity. In this region, 2 previously unreported IL6ST variants, p.K702Sfs7* and p.Y759Wfs26*, were identified in 2 families with autosomal dominant (AD) HIES. Both variants were LOF and exhibited DN effects, leading to the accumulation of mutant GP130 on the cell surface. The p.K702Sfs7* mutation was the most upstream N-terminal mutation linked to HIES caused by heterozygous IL6ST variants. Comprehensive screening of IL6ST mutants revealed that most premature terminations downstream of amino acid F641, at the end of the transmembrane domain, resulted in LOF and DN effects via GP130 accumulation on the cell surface. The absence of the recycling motif (positions 782-787) in surface-expressed LOF GP130 led to its accumulation, contributing to the DN effect. The importance of intracellular truncating IL6ST variants can possibly be predicted based on the location of the premature stop codon. GP130 accumulation on the cell surface is a characteristic and potentially diagnostic finding in patients with HIES with heterozygous IL6ST variants.
Product Citations: 67
In JCI Insight on 22 July 2025 by Ashihara, K., Asano, T., et al.
-
FC/FACS
-
Homo sapiens (Human)
Preprint on BioRxiv : the Preprint Server for Biology on 4 February 2025 by Castelli, S., Wilson, W. V., et al.
Summary The success of chimeric antigen receptor T cell therapies targeting solid tumors is limited by the immunosuppressive tumor microenvironment. We demonstrate that endowing CAR T cells with ectopic interleukin-9 (IL-9) signaling by co-expressing an IL-9 receptor, rewires CAR T cell fate under antigen stress to enhance anti-tumor efficacy. In preclinical solid tumor models, IL-9-signaling CAR T cells exhibit increased expansion, persistence, and tumor infiltration, resulting in superior tumor control at significantly lower doses than conventional products. Trajectory and RNA velocity analyses of single-cell RNA sequencing data reveal that IL-9 signaling alters CAR T cell differentiation under antigen stress away from dysfunction, favoring a multipotent transition toward CD8+ cell memory and effector states, and promoting a CD4+ cell proliferative state. Interrogation of transcription factor pathways indicates that IL-9-mediated activation of STAT1 and STAT4 drives the superior phenotype of IL-9-signaling CAR T cells, providing a promising therapeutic strategy for targeting solid cancers.
-
Cancer Research
-
Immunology and Microbiology
-
Stem Cells and Developmental Biology
In Molecular Therapy. Nucleic Acids on 10 September 2024 by Wang, D., Kaniowski, D., et al.
Acute myeloid leukemia (AML) cells resist differentiation stimuli despite high expression of innate immune receptors, such as Toll-like receptor 9 (TLR9). We previously demonstrated that targeting Signal Transducer and Activator of Transcription 3 (STAT3) using TLR9-targeted decoy oligodeoxynucleotide (CpG-STAT3d) increases immunogenicity of human and mouse AML cells. Here, we elucidated molecular mechanisms of inv(16) AML reprogramming driven by STAT3-inhibition/TLR9-activation in vivo. At the transcriptional levels, AML cells isolated from mice after intravenous administration of CpG-STAT3d or leukemia-targeted Stat3 silencing and TLR9 co-stimulation, displayed similar upregulation of myeloid cell differentiation (Irf8, Cebpa, Itgam) and antigen-presentation (Ciita, Il12a, B2m)-related genes with concomitant reduction of leukemia-promoting Runx1. Single-cell transcriptomics revealed that CpG-STAT3d induced multilineage differentiation of AML cells into monocytes/macrophages, erythroblastic and B cell subsets. As shown by an inducible Irf8 silencing in vivo, IRF8 upregulation was critical for monocyte-macrophage differentiation of leukemic cells. TLR9-driven AML cell reprogramming was likely enabled by downregulation of STAT3-controlled methylation regulators, such as DNMT1 and DNMT3. In fact, the combination of DNA methyl transferase (DNMT) inhibition using azacitidine with CpG oligonucleotides alone mimicked CpG-STAT3d effects, resulting in AML cell differentiation, T cell activation, and systemic leukemia regression. These findings highlight immunotherapeutic potential of bi-functional oligonucleotides to unleash TLR9-driven differentiation of leukemic cells by concurrent STAT3 and/or DNMT inhibition.
© 2024 The Author(s).
-
Mus musculus (House mouse)
-
Cancer Research
Improving Reliability of Immunological Assays by Defining Minimal Criteria for Cell Fitness.
In ImmunoHorizons on 1 September 2024 by Ivison, S., Boucher, G., et al.
Human PBMC-based assays are often used as biomarkers for the diagnosis and prognosis of disease, as well as for the prediction and tracking of response to biological therapeutics. However, the development and use of PBMC-based biomarker assays is often limited by poor reproducibility. Complex immunological assays can be further complicated by variation in cell handling before analysis, especially when using cryopreserved cells. Variation in postthaw viability is further increased if PBMC isolation and cryopreservation are done more than a few hours after collection. There is currently a lack of evidence-based standards for the minimal PBMC viability or "fitness" required to ensure the integrity and reproducibility of immune cell-based assays. In this study, we use an "induced fail" approach to examine the effect of thawed human PBMC fitness on four flow cytometry-based assays. We found that cell permeability-based viability stains at the time of thawing did not accurately quantify cell fitness, whereas a combined measurement of metabolic activity and early apoptosis markers did. Investigation of the impact of different types and levels of damage on PBMC-based assays revealed that only when cells were >60-70% live and apoptosis negative did biomarker values cease to be determined by cell fitness rather than the inherent biology of the cells. These data show that, to reproducibly measure immunological biomarkers using cryopreserved PBMCs, minimal acceptable standards for cell fitness should be incorporated into the assay protocol.
Copyright © 2024 The Authors.
-
Immunology and Microbiology
Preclinical proof of principle for orally delivered Th17 antagonist miniproteins.
In Cell on 8 August 2024 by Berger, S., Seeger, F., et al.
Interleukin (IL)-23 and IL-17 are well-validated therapeutic targets in autoinflammatory diseases. Antibodies targeting IL-23 and IL-17 have shown clinical efficacy but are limited by high costs, safety risks, lack of sustained efficacy, and poor patient convenience as they require parenteral administration. Here, we present designed miniproteins inhibiting IL-23R and IL-17 with antibody-like, low picomolar affinities at a fraction of the molecular size. The minibinders potently block cell signaling in vitro and are extremely stable, enabling oral administration and low-cost manufacturing. The orally administered IL-23R minibinder shows efficacy better than a clinical anti-IL-23 antibody in mouse colitis and has a favorable pharmacokinetics (PK) and biodistribution profile in rats. This work demonstrates that orally administered de novo-designed minibinders can reach a therapeutic target past the gut epithelial barrier. With high potency, gut stability, and straightforward manufacturability, de novo-designed minibinders are a promising modality for oral biologics.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
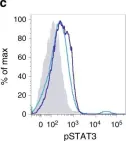
In Nat Commun on 3 December 2018 by Calcinotto, A., Brevi, A., et al.
Fig.3.C

-
FC/FACS
-
Mus musculus (House mouse)
Collected and cropped from Nat Commun by CiteAb, provided under a CC-BY license
Image 1 of 2
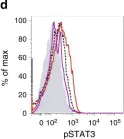
In Nat Commun on 3 December 2018 by Calcinotto, A., Brevi, A., et al.
Fig.3.D

-
FC/FACS
-
Mus musculus (House mouse)
Collected and cropped from Nat Commun by CiteAb, provided under a CC-BY license
Image 1 of 2