The ENCODE Consortium's efforts to annotate noncoding cis-regulatory elements (CREs) have advanced our understanding of gene regulatory landscapes. Pooled, noncoding CRISPR screens offer a systematic approach to investigate cis-regulatory mechanisms. The ENCODE4 Functional Characterization Centers conducted 108 screens in human cell lines, comprising >540,000 perturbations across 24.85 megabases of the genome. Using 332 functionally confirmed CRE-gene links in K562 cells, we established guidelines for screening endogenous noncoding elements with CRISPR interference (CRISPRi), including accurate detection of CREs that exhibit variable, often low, transcriptional effects. Benchmarking five screen analysis tools, we find that CASA produces the most conservative CRE calls and is robust to artifacts of low-specificity single guide RNAs. We uncover a subtle DNA strand bias for CRISPRi in transcribed regions with implications for screen design and analysis. Together, we provide an accessible data resource, predesigned single guide RNAs for targeting 3,275,697 ENCODE SCREEN candidate CREs with CRISPRi and screening guidelines to accelerate functional characterization of the noncoding genome.
© 2024. The Author(s).
Product Citations: 10
Multicenter integrated analysis of noncoding CRISPRi screens.
In Nature Methods on 1 April 2024 by Yao, D., Tycko, J., et al.
Guidelines for the use of flow cytometry and cell sorting in immunological studies (third edition).
In European Journal of Immunology on 1 December 2021 by Cossarizza, A., Chang, H. D., et al.
The third edition of Flow Cytometry Guidelines provides the key aspects to consider when performing flow cytometry experiments and includes comprehensive sections describing phenotypes and functional assays of all major human and murine immune cell subsets. Notably, the Guidelines contain helpful tables highlighting phenotypes and key differences between human and murine cells. Another useful feature of this edition is the flow cytometry analysis of clinical samples with examples of flow cytometry applications in the context of autoimmune diseases, cancers as well as acute and chronic infectious diseases. Furthermore, there are sections detailing tips, tricks and pitfalls to avoid. All sections are written and peer-reviewed by leading flow cytometry experts and immunologists, making this edition an essential and state-of-the-art handbook for basic and clinical researchers.
© 2021 Wiley-VCH GmbH.
-
FC/FACS
-
Mus musculus (House mouse)
-
Immunology and Microbiology
In Immunity on 15 September 2020 by Kim, D., Nguyen, Q. T., et al.
Glucocorticoids (GC) are the mainstay treatment option for inflammatory conditions. Despite the broad usage of GC, the mechanisms by which GC exerts its effects remain elusive. Here, utilizing murine autoimmune and allergic inflammation models, we report that Foxp3+ regulatory T (Treg) cells are irreplaceable GC target cells in vivo. Dexamethasone (Dex) administered in the absence of Treg cells completely lost its ability to control inflammation, and the lack of glucocorticoid receptor in Treg cells alone resulted in the loss of therapeutic ability of Dex. Mechanistically, Dex induced miR-342-3p specifically in Treg cells and miR-342-3p directly targeted the mTORC2 component, Rictor. Altering miRNA-342-3p or Rictor expression in Treg cells dysregulated metabolic programming in Treg cells, controlling their regulatory functions in vivo. Our results uncover a previously unknown contribution of Treg cells during glucocorticoid-mediated treatment of inflammation and the underlying mechanisms operated via the Dex-miR-342-Rictor axis.
Copyright © 2020 Elsevier Inc. All rights reserved.
-
Mus musculus (House mouse)
-
Immunology and Microbiology
Cancer-Specific Loss of p53 Leads to a Modulation of Myeloid and T Cell Responses.
In Cell Reports on 14 January 2020 by Blagih, J., Zani, F., et al.
Loss of p53 function contributes to the development of many cancers. While cell-autonomous consequences of p53 mutation have been studied extensively, the role of p53 in regulating the anti-tumor immune response is still poorly understood. Here, we show that loss of p53 in cancer cells modulates the tumor-immune landscape to circumvent immune destruction. Deletion of p53 promotes the recruitment and instruction of suppressive myeloid CD11b+ cells, in part through increased expression of CXCR3/CCR2-associated chemokines and macrophage colony-stimulating factor (M-CSF), and attenuates the CD4+ T helper 1 (Th1) and CD8+ T cell responses in vivo. p53-null tumors also show an accumulation of suppressive regulatory T (Treg) cells. Finally, we show that two key drivers of tumorigenesis, activation of KRAS and deletion of p53, cooperate to promote immune tolerance.Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
-
Mus musculus (House mouse)
-
Cancer Research
-
Immunology and Microbiology
In Mucosal Immunology on 1 November 2019 by Gribonika, I., Eliasson, D. G., et al.
Our understanding of how class-switch recombination (CSR) to IgA occurs in the gut is still incomplete. Earlier studies have indicated that Tregs are important for IgA CSR and these cells were thought to transform into follicular helper T cells (Tfh), responsible for germinal center formation in the Peyer's patches (PP). Following adoptive transfer of T-cell receptor-transgenic (TCR-Tg) CD4 T cells into nude mice, we unexpectedly found that oral immunization did not require an adjuvant to induce strong gut IgA and systemic IgG responses, suggesting an altered regulatory environment in the PP. After sorting of splenic TCR-Tg CD4 T cells into CD25+ or CD25- cells we observed that none of these fractions supported a gut IgA response, while IgG responses were unperturbed in mice receiving the CD25- cell fraction. Hence, while Tfh functions resided in the CD25- fraction the IgA CSR function in the PP was dependent on CD25+ Foxp3+ Tregs, which were found to be Helios+ neuropilin-1+ thymus-derived Tregs. This is the first study to demonstrate that Tfh and IgA CSR functions are indeed, unique, and separate functions in the PP with the former being TCR-dependent while the latter appeared to be antigen independent.
-
Immunology and Microbiology
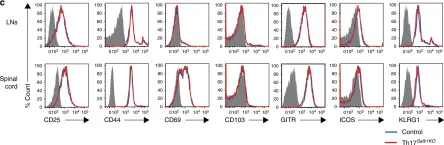
In Nat Commun on 1 February 2019 by Yasuda, K., Kitagawa, Y., et al.
Fig.3.C

-
FC/FACS
-
Mus musculus (House mouse)
Collected and cropped from Nat Commun by CiteAb, provided under a CC-BY license
Image 1 of 1