Myelodysplastic syndrome disease (MDS) is caused by the successive acquisition of mutations and thus displays a variable risk for progression to AML. Mutations in CEBPA are commonly associated with a high risk of disease progression, but whether they are causative for AML development is unclear. To analyse the molecular basis of disease progression we generated MDS patient-derived induced pluripotent stem cells from a low risk male patient harbouring RUNX1/SRSF2 mutations. This experimental model faithfully recapitulates the patient disease phenotypes upon hematopoietic differentiation. Introduction of a frameshift mutation affecting the C/EBPα bZIP domain in cells from low-risk stages mimicks disease progression by reducing clonogenicity of myeloid cells, blocking granulopoiesis and increasing erythroid progenitor self-renewal capacity. The acquisition of this mutation reshapes the chromatin landscape at distal cis-regulatory regions and promotes changes in cellular composition as observed by single cell RNAseq. Mutant C/EBPα is therefore causative for MDS disease progression. Our work identifies mutant CEBPA as causative for MDS disease progression, providing a new isogenic MDS experimental model for drug screening to improve diagnostic and therapeutic strategies.
© 2025. The Author(s).
Product Citations: 36
In Nature Communications on 1 July 2025 by Almaghrabi, R., Alyahyawi, Y., et al.
-
FC/FACS
-
Homo sapiens (Human)
Preprint on Research Square on 4 September 2024 by Giorgetti, A., Marin-Béjar, O., et al.
Abstract Patients with GATA2 deficiency are predisposed to developing myelodysplastic syndrome (MDS), which can progress to acute myeloid leukemia (AML). This progression is often associated with the acquisition of additional cytogenetic and somatic alterations. Mutations in SETBP1 and ASXL1 genes are frequently observed in pediatric GATA2 patients, but their roles in disease progression remain poorly understood. Genome editing of induced pluripotent stem cells (iPSCs) enabled precise reconstruction of mutation combinations found in patients. Here we developed a human hiPSC-based model to study the impact of SETBP1 and ASXL1 mutations in context of GATA2 deficiency. We show that germline heterozygous GATA2 mutation alone showed no significant effect on myeloid development, while the addition of SETBP1 and ASXL1 mutations impaired myelopoiesis, resulting in monocytopenia. We identified a key role of the SETBP1 mutation in promoting chromatin remodeling near genes involved in myeloid neoplasms, which likely initiated the blockage of myeloid differentiation observed in vitro. Motif analysis of more accessible chromatin regions in the SETBP1 and SETBP1/ASXL1 mutant background highlighted an enrichment for MEIS1, PU.1, RUNX1, and HOXA9, implicating these factors in the disease progression. Our study establishes a novel humanized model system for studying GATA2 deficiency. We demonstrate that SETBP1 mutations act as a primary driver in hematopoietic impairment, providing insights that may inform future therapeutic strategies for patients progressing to MDS/AML
-
Homo sapiens (Human)
In Developmental Cell on 6 May 2024 by Fowler, J. L., Zheng, S. L., et al.
The developmental origin of blood-forming hematopoietic stem cells (HSCs) is a longstanding question. Here, our non-invasive genetic lineage tracing in mouse embryos pinpoints that artery endothelial cells generate HSCs. Arteries are transiently competent to generate HSCs for 2.5 days (∼E8.5-E11) but subsequently cease, delimiting a narrow time frame for HSC formation in vivo. Guided by the arterial origins of blood, we efficiently and rapidly differentiate human pluripotent stem cells (hPSCs) into posterior primitive streak, lateral mesoderm, artery endothelium, hemogenic endothelium, and >90% pure hematopoietic progenitors within 10 days. hPSC-derived hematopoietic progenitors generate T, B, NK, erythroid, and myeloid cells in vitro and, critically, express hallmark HSC transcription factors HLF and HOXA5-HOXA10, which were previously challenging to upregulate. We differentiated hPSCs into highly enriched HLF+ HOXA+ hematopoietic progenitors with near-stoichiometric efficiency by blocking formation of unwanted lineages at each differentiation step. hPSC-derived HLF+ HOXA+ hematopoietic progenitors could avail both basic research and cellular therapies.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
-
FC/FACS
-
Homo sapiens (Human)
-
Stem Cells and Developmental Biology
Protocol for differentiation of functional macrophages from human induced pluripotent stem cells.
In STAR Protocols on 15 March 2024 by Jeong, S., Chang, H., et al.
Human induced pluripotent stem cell (hiPSC)-derived macrophages provide a valuable tool for disease modeling and drug discovery. Here, we present a protocol to generate functional macrophages from hiPSCs using a feeder-free hematopoietic differentiation technique. We describe steps for preparing hiPSCs, mesodermal differentiation, hematopoietic commitment, and macrophage differentiation and expansion. We then detail assays to characterize their phenotype, polarization, and phagocytic functions. The functional macrophages generated here could be used to generate organoids for disease modeling and drug discovery studies. For complete details on the use and execution of this protocol, please refer to Jeong et al.1 and Heo et al.2.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
-
Stem Cells and Developmental Biology
In Immunooncol Technol on 1 December 2023 by Jin, G., Chang, Y., et al.
Glioblastoma (GBM) is an aggressive brain tumor giving a poor prognosis with the current treatment options. The advent of chimeric antigen receptor (CAR) T-cell therapy revolutionized the field of immunotherapy and has provided a new set of therapeutic options for refractory blood cancers. In an effort to apply this therapeutic approach to solid tumors, various immune cell types and CAR constructs are being studied. Notably, macrophages have recently emerged as potential candidates for targeting solid tumors, attributed to their inherent tumor-infiltrating capacity and abundant presence in the tumor microenvironment.
In this study, we developed a chemically defined differentiation protocol to generate macrophages from human pluripotent stem cells (hPSCs). A GBM-specific CAR was genetically incorporated into hPSCs to generate CAR hPSC-derived macrophages.
The CAR hPSC-derived macrophages exhibited potent anticancer activity against GBM cells in vitro.
Our findings demonstrate the feasibility of generating functional CAR-macrophages from hPSCs for adoptive immunotherapy, thereby opening new avenues for the treatment of solid tumors, particularly GBM.
© 2023 The Author(s).
-
FC/FACS
-
Immunology and Microbiology
-
Stem Cells and Developmental Biology
In PLoS One on 16 December 2020 by Dubois, F., Gaignerie, A., et al.
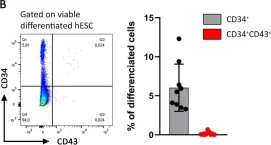
Fig.1.B

-
FC/FACS
-
Collected and cropped from PLoS One by CiteAb, provided under a CC-BY license
Image 1 of 1