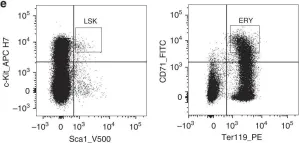
Alterations in DNA methylation occur during development, but the mechanisms by which they influence gene expression remain uncertain. There are few examples where modification of a single CpG dinucleotide directly affects transcription factor binding and regulation of a target gene in vivo. Here, we show that the erythroid transcription factor GATA-1 - that typically binds T/AGATA sites - can also recognise CGATA elements, but only if the CpG dinucleotide is unmethylated. We focus on a single CGATA site in the c-Kit gene which progressively becomes unmethylated during haematopoiesis. We observe that methylation attenuates GATA-1 binding and gene regulation in cell lines. In mice, converting the CGATA element to a TGATA site that cannot be methylated leads to accumulation of megakaryocyte-erythroid progenitors. Thus, the CpG dinucleotide is essential for normal erythropoiesis and this study illustrates how a single methylated CpG can directly affect transcription factor binding and cellular regulation.
Product Citations: 4
Methylation of a CGATA element inhibits binding and regulation by GATA-1.
In Nature Communications on 22 May 2020 by Yang, L., Chen, Z., et al.
-
FC/FACS
-
Mus musculus (House mouse)
In Cell Reports on 20 November 2018 by Nakagawa, M. M. & Rathinam, C. V.
Constitutive activation of the canonical NF-κB pathway has been associated with a variety of human pathologies. However, molecular mechanisms through which canonical NF-κB affects hematopoiesis remain elusive. Here, we demonstrate that deregulated canonical NF-κB signals in hematopoietic stem cells (HSCs) cause a complete depletion of HSC pool, pancytopenia, bone marrow failure, and premature death. Constitutive activation of IKK2 in HSCs leads to impaired quiescence and loss of function. Gene set enrichment analysis (GSEA) identified an induction of "erythroid signature" in HSCs with augmented NF-κB activity. Mechanistic studies indicated a reduction of thrombopoietin (TPO)-mediated signals and its downstream target p57 in HSCs, due to reduced c-Mpl expression in a cell-intrinsic manner. Molecular studies established Klf1 as a key suppressor of c-Mpl in HSPCs with increased NF-κB. In essence, these studies identified a previously unknown mechanism through which exaggerated canonical NF-κB signals affect HSCs and cause pathophysiology.
Published by Elsevier Inc.
-
FC/FACS
-
Mus musculus (House mouse)
-
Stem Cells and Developmental Biology
Persistent response of Fanconi anemia haematopoietic stem and progenitor cells to oxidative stress.
In Cell Cycle on 18 June 2017 by Li, Y., Amarachintha, S., et al.
Oxidative stress is considered as an important pathogenic factor in many human diseases including Fanconi anemia (FA), an inherited bone marrow failure syndrome with extremely high risk of leukemic transformation. Members of the FA protein family are involved in DNA damage and other cellular stress responses. Loss of FA proteins renders cells hypersensitive to oxidative stress and cancer transformation. However, how FA cells respond to oxidative DNA damage remains unclear. By using an in vivo stress-response mouse strain expressing the Gadd45β-luciferase transgene, we show here that haematopoietic stem and progenitor cells (HSPCs) from mice deficient for the FA gene Fanca or Fancc persistently responded to oxidative stress. Mechanistically, we demonstrated that accumulation of unrepaired DNA damage, particularly in oxidative damage-sensitive genes, was responsible for the long-lasting response in FA HSPCs. Furthermore, genetic correction of Fanca deficiency almost completely abolished the persistent oxidative stress-induced G2/M arrest and DNA damage response in vivo. Our study suggests that FA pathway is an integral part of a versatile cellular mechanism by which HSPCs respond to oxidative stress.
-
FC/FACS
-
Mus musculus (House mouse)
-
Cell Biology
In Cell Reports on 27 October 2015 by Morgado-Palacin, L., Varetti, G., et al.
Diamond-Blackfan anemia (DBA) is characterized by anemia and cancer susceptibility and is caused by mutations in ribosomal genes, including RPL11. Here, we report that Rpl11-heterozygous mouse embryos are not viable and that Rpl11 homozygous deletion in adult mice results in death within a few weeks, accompanied by bone marrow aplasia and intestinal atrophy. Importantly, Rpl11 heterozygous deletion in adult mice results in anemia associated with decreased erythroid progenitors and defective erythroid maturation. These defects are also present in mice transplanted with inducible heterozygous Rpl11 bone marrow and, therefore, are intrinsic to the hematopoietic system. Additionally, heterozygous Rpl11 mice present increased susceptibility to radiation-induced lymphomagenesis. In this regard, total or partial deletion of Rpl11 compromises p53 activation upon ribosomal stress or DNA damage in fibroblasts. Moreover, fibroblasts and hematopoietic tissues from heterozygous Rpl11 mice present higher basal cMYC levels. We conclude that Rpl11-deficient mice recapitulate DBA disorder, including cancer predisposition.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
-
FC/FACS
-
Mus musculus (House mouse)
In Nat Commun on 22 May 2020 by Yang, L., Chen, Z., et al.
Fig.2.E

-
FC/FACS
-
Mus musculus (House mouse)
Collected and cropped from Nat Commun by CiteAb, provided under a CC-BY license
Image 1 of 1