Alpha-kinase 1 (ALPK1) is an immune receptor sensing the bacterial nucleotide sugar ADP-heptose. ALPK1 phosphorylates TIFA leading to its oligomerization and downstream NF-κB activation. Specific mutations in ALPK1 are associated with an autoinflammatory syndrome termed ROSAH and with spiradenoma (skin cancers with sweat gland differentiation). This study investigated ALPK1 responses in human mononuclear cells and demonstrates that human mononuclear cells have distinct abilities to respond to ADP-heptose. Notably, IFN-γ is required to license the ALPK1/TIFA pathway in monocytes, while it was dispensable for the responsiveness of B cells. IFN-γ induced TIFA upregulation in monocytes, and TIFA induction was sufficient to recapitulate the licensing effect of IFN-γ. IFN-γ treatment promoted the phenotypic expression of pathogenic ALPK1 mutations. The licensing effect of IFN-γ in monocytes was blocked by JAK inhibitors. These findings underscore the critical role of IFN-γ in ALPK1 function and suggest JAK inhibitors as potential therapies for ALPK1-related inflammatory conditions.
© 2024 The Author(s).
Product Citations: 64
IFN-γ licenses normal and pathogenic ALPK1/TIFA pathway in human monocytes.
In IScience on 17 January 2025 by Martin, A., Caron, S., et al.
Single-cell analysis reveals islet autoantigen's immune activation in type 1 diabetes patients.
In Journal of Clinical Biochemistry and Nutrition on 1 January 2025 by Okamura, T., Kitagawa, N., et al.
In this study, we used single-cell sequencing, which can comprehensively detect the type and number of transcripts per cell, to efficiently stimulate peripheral blood mononuclear cells of type 1 diabetic patients with overlapping peptides of GAD, IA-2, and insulin antigens, and performed gene expression analysis by single-cell variable-diversity-joining sequencing and T-cell receptor repertoire analysis. Twenty male patients with type 1 diabetes mellitus participating in the KAMOGAWA-DM cohort were included. Four of them were randomly selected for BD Rhapsody system after reacting peripheral blood mononuclear cells with overlapping peptides of GAD, IA-2, and insulin antigen. Peripheral blood mononuclear cells were clustered into CD8+ T cells, CD4+ T cells, granulocytes, natural killer cells, dendritic cells, monocytes, and B cells based on Seurat analysis. In the insulin group, gene expression of inflammatory cytokines was elevated in cytotoxic CD8+ T cells and Th1 and Th17 cells, and gene expression related to exhaustion was elevated in regulatory T cells. In T cell receptors of various T cells, the T cell receptor β chain was monoclonally increased in the TRBV28/TRBJ2-7 pairs. This study provides insights into the pathogenesis of type 1 diabetes and provides potential targets for the treatment of type 1 diabetes.
Copyright © 2025 JCBN.
-
Homo sapiens (Human)
-
Biochemistry and Molecular biology
-
Immunology and Microbiology
Tuberculosis in otherwise healthy adults with inherited TNF deficiency.
In Nature on 1 September 2024 by Arias, A. A., Neehus, A. L., et al.
Severe defects in human IFNγ immunity predispose individuals to both Bacillus Calmette-Guérin disease and tuberculosis, whereas milder defects predispose only to tuberculosis1. Here we report two adults with recurrent pulmonary tuberculosis who are homozygous for a private loss-of-function TNF variant. Neither has any other clinical phenotype and both mount normal clinical and biological inflammatory responses. Their leukocytes, including monocytes and monocyte-derived macrophages (MDMs) do not produce TNF, even after stimulation with IFNγ. Blood leukocyte subset development is normal in these patients. However, an impairment in the respiratory burst was observed in granulocyte-macrophage colony-stimulating factor (GM-CSF)-matured MDMs and alveolar macrophage-like (AML) cells2 from both patients with TNF deficiency, TNF- or TNFR1-deficient induced pluripotent stem (iPS)-cell-derived GM-CSF-matured macrophages, and healthy control MDMs and AML cells differentiated with TNF blockers in vitro, and in lung macrophages treated with TNF blockers ex vivo. The stimulation of TNF-deficient iPS-cell-derived macrophages with TNF rescued the respiratory burst. These findings contrast with those for patients with inherited complete deficiency of the respiratory burst across all phagocytes, who are prone to multiple infections, including both Bacillus Calmette-Guérin disease and tuberculosis3. Human TNF is required for respiratory-burst-dependent immunity to Mycobacterium tuberculosis in macrophages but is surprisingly redundant otherwise, including for inflammation and immunity to weakly virulent mycobacteria and many other infectious agents.
© 2024. The Author(s).
Use of human AML cells to study graft-versus-leukemia immunity in xenogeneic mouse models of GVHD
Preprint on BioRxiv : the Preprint Server for Biology on 2 May 2024 by Faville, C., Silva, B. E., et al.
Allogeneic hematopoietic cell transplantation (allo-HCT) is the main therapeutic approach for patients with high-risk acute myeloid leukemia (AML), but the rate of relapse remains high and is associated with poor outcomes. Discovering new approaches to maximize the graft-versus-leukemia (GVL) effects while mitigating graft-versus-host disease (GVHD) should therefore be pursued. Because of the difficulties in modeling AML in mice, patient-derived xenotransplantations (PDX) in immunodeficient NSG mice are preferred to study the GVL effects. In PDX, AML is typically induced through the intravenous injection of cell lines or leukemic blasts obtained from patients. GVHD and GVL effects are induced by (co)-injecting human T cells or peripheral blood mononuclear cells (PBMCs). While this approach enables the induction of systemic leukemia, notably developing in the spleen and bone marrow of the animals, it can also be associated with difficulties in monitoring the disease, notably by flow cytometry. This can be circumvented by using luciferase-expressing AML cells or transplanting the leukemic cells in Matrigel to generate solid tumors that are easier to monitor. Here, we provide detailed instructions on how to prepare human PBMCs and leukemic cells, transplant them, and monitor the disease in NSG mice.
-
Cancer Research
-
Immunology and Microbiology
In Methods in Molecular Biology (Clifton, N.J.) on 16 April 2024 by do Prado Servian, C., Masson, L. C., et al.
The encounter of T cells with the antigen through the interaction of T cell receptors with peptides and major histocompatibility complex (MHC) molecules on the surface of antigen-presenting cells (APCs) can generate effector response and memory T cells. Memory T cells developed following infections or vaccination may persist, leading to the generation of a specific immune response upon reexposure to the same pathogen through rapid clonal proliferation and activation of effector functions. T cell memory subsets can be identified based on the expression of several membrane markers such as CCR7, CD27, and CD45RA. Using fluorescent antibodies against these markers and a flow cytometer, it is possible to perform immunophenotyping via the analysis of cell surface expression of proteins by different subpopulations such as the subsets of naïve, effector, and memory T cells as well as via the analysis of functional markers that further characterize each sample. Intracellular cytokine staining allows for the evaluation of intracellular proteins expressed in T cells in response to antigenic stimulation. This chapter presents the phenotypic and functional characterization of memory T cells after antigenic stimulation, detailing the procedures for identifying intracellular and surface protein markers. Herein, we review and present a reproducible standardized protocol using antibodies for specific markers and applying flow cytometry.
© 2024. The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature.
-
Biochemistry and Molecular biology
-
Immunology and Microbiology
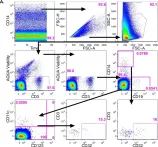
In PLoS One on 11 May 2016 by Cheeseman, H. M., Carias, A. M., et al.
Fig.2.A

-
FC/FACS
-
Homo sapiens (Human)
Collected and cropped from PLoS One by CiteAb, provided under a CC-BY license
Image 1 of 2
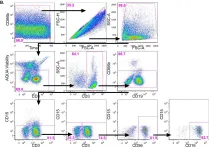
In PLoS One on 11 May 2016 by Cheeseman, H. M., Carias, A. M., et al.
Fig.2.B

-
FC/FACS
-
Homo sapiens (Human)
Collected and cropped from PLoS One by CiteAb, provided under a CC-BY license
Image 1 of 2