One of the challenges in Good Manufacturing Practice (GMP)-compliant human induced pluripotent stem cell (hiPSC) production is the validation of quality control (QC) tests specific for hiPSCs, which are required for GMP batch release. This study presents a comprehensive description of the validation process for hiPSC-specific GMP-compliant QC assays; more specifically, the validation of assays to assess the potential presence of residual episomal vectors (REVs), the expression of markers of the undifferentiated state and the directed differentiation potential of hiPSCs. Critical aspects and specific acceptance criteria were formulated in a validation plan prior to assay validation. Assay specificity, sensitivity and reproducibility were tested, and the equipment used for each assay was subjected to performance qualification. A minimum input of 20 000 cells (120 ng of genomic DNA) was defined for accurate determination of the presence of REVs. Furthermore, since vector loss in hiPSC lines is a passage-dependent process, we advocate screening for REVs between passages eight and 10, as testing at earlier passages might lead to unnecessary rejection of hiPSC lines. The cutoff value for assessment of markers of the undifferentiated state was set to the expression of at least three individual markers on at least 75% of the cells. When multi-color flow cytometry panels are used, a fluorescence minus one control is advised to ensure the control for fluorescent spread. For the assay to assess the directed differentiation potential, the detection limit was set to two of three positive lineage-specific markers for each of the three individual germ layers. All of our assays proved to be reproducible and specific. Our data demonstrate that our implemented analytical procedures are suitable as QC assays for the batch release of GMP-compliant hiPSCs.
Copyright © 2024 International Society for Cell & Gene Therapy. Published by Elsevier Inc. All rights reserved.
Product Citations: 27
In Cytotherapy on 1 September 2024 by Novoa, J., Westra, I., et al.
-
Homo sapiens (Human)
-
Stem Cells and Developmental Biology
In Stem Cell Research on 1 March 2023 by Arendzen, C. H., Cramer, S. J., et al.
We previously generated a doxycycline-inducible H2B-mTurq2 reporter in hiPSCs to track cells and study cell division and apoptosis. To improve visualization of cycling cells, we introduced a ubiquitously transcribed mScarletI-Geminin (GMMN) (1-110) into the previously untargeted second AAVS1 allele. Fusion to the N-terminal part of GMNN provided tightly controlled mScarletI expression during the cell cycle. mScarletI fluorescence increased gradually from the S-phase through the M-phase of the cell cycle and was lost at the metaphase-anaphase transition. The resulting hiPSC reporter line generated, which we named ProLiving, is a valuable tool to study cell division and cell cycle characteristics in living hiPSC-derived cells.
Copyright © 2023 The Authors. Published by Elsevier B.V. All rights reserved.
-
Homo sapiens (Human)
-
Stem Cells and Developmental Biology
Generation of AAVS1 and CLYBL STRAIGHT-IN v2 acceptor human iPSC lines for integrating DNA payloads.
In Stem Cell Research on 1 February 2023 by Blanch-Asensio, A., van der Vaart, B., et al.
STRAIGHT-IN is a platform to precisely integrate DNA payloads into the genome of cells, including hiPSCs. Here, we generated two hiPSC acceptor lines each with one copy of an upgraded landing pad (LP). This improved design allows more efficient (∼100 %) and rapid (∼2-3 weeks) generation of genetically modified hiPSC lines containing the desired payloads. This new LP version was inserted into either the AAVS1 (LUMCi004-A-1) or CLYBL (LUMCi004-A-2) safe harbour loci in the hiPSC line, LUMC0099iCTRL04. The resulting lines can be used for the targeted integration of a wide range of transgenes, thereby making them suitable for numerous research applications.
Copyright © 2022 The Authors. Published by Elsevier B.V. All rights reserved.
-
Homo sapiens (Human)
-
Genetics
-
Stem Cells and Developmental Biology
In Stem Cells Translational Medicine on 18 November 2022 by Hekman, K. E., Koss, K. M., et al.
Stem cells are enabling an improved understanding of the peripheral arterial disease, and patient-specific stem cell-derived endothelial cells (ECs) present major advantages as a therapeutic modality. However, applications of patient-specific induced pluripotent stem cell (iPSC)-derived ECs are limited by rapid loss of mature cellular function in culture. We hypothesized that changes in autophagy impact the phenotype and cellular proliferation of iPSC-ECs. Endothelial cells were differentiated from distinct induced pluripotent stem cell lines in 2D culture and purified for CD144 positive cells. Autophagy, mitochondrial morphology, and proliferation were characterized during differentiation and over serial passages in culture. We found that autophagy activity was stimulated during differentiation but stagnated in mature iPSC-ECs. Mitochondria remodeled through mitophagy during differentiation and demonstrated increasing membrane potential and mass through serial passages; however, these plateaued, coinciding with decreased proliferation. To evaluate for oxidative damage, iPSC-ECs were alternatively grown under hypoxic culture conditions; however, hypoxia only transiently improved the proliferation. Stimulating mTOR-independent ULK1-mediated autophagy with a plant derivative AMP kinase activator Rg2 significantly improved proliferative capacity of iPSC-ECs over multiple passages. Therefore, autophagy, a known mediator of longevity, played an active role in remodeling mitochondria during maturation from pluripotency to a terminally differentiated state. Autophagy failed to compensate for increasing mitochondrial mass over serial passages, which correlated with loss of proliferation in iPSC-ECs. Stimulating ULK1-kinase-driven autophagy conferred improved proliferation and longevity over multiple passages in culture. This represents a novel approach to overcoming a major barrier limiting the use of iPSC-ECs for clinical and research applications.
© The Author(s) 2022. Published by Oxford University Press.
-
FC/FACS
-
Cell Biology
-
Stem Cells and Developmental Biology
In Stem Cell Research on 1 December 2021 by Arendzen, C. H., Chaudhari, U., et al.
An induced pluripotent stem cell (iPSC) line, in which a H2B-fluorescent protein fusion is temporally expressed, is a valuable tool to track cells and study cell divisions and apoptosis. To this end we introduced a 3rd generation "all-in-one" doxycycline-inducible H2B-mTurquoise2 vector into the AAVS1 locus of PAX3-Venus iPSCs via CRISPR/Cas9. H2B-mTurquoise2 expression is absent but readily induced by doxycycline allowing quantification of cell divisions and imaging of living cells. Besides being a universal reporter in iPSC-based differentiation and toxicity assays, the generated pluripotent and genomically normal LUMCi041-A-2 line is particularly suited to study PAX3-positive stages of development.
Copyright © 2021 The Authors. Published by Elsevier B.V. All rights reserved.
-
Homo sapiens (Human)
-
Stem Cells and Developmental Biology
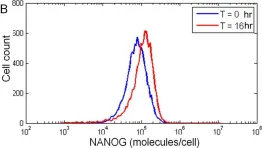
In PLoS One on 12 December 2012 by Wu, J. & Tzanakakis, E. S.
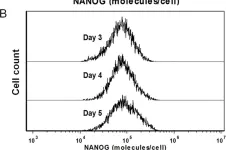
Fig.2.B

-
FC/FACS
-
Collected and cropped from PLoS One by CiteAb, provided under a CC-BY license
Image 1 of 7
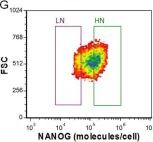
In PLoS One on 12 December 2012 by Wu, J. & Tzanakakis, E. S.
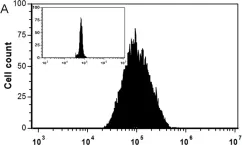
Fig.2.A

-
FC/FACS
-
Collected and cropped from PLoS One by CiteAb, provided under a CC-BY license
Image 1 of 7
In PLoS One on 12 December 2012 by Wu, J. & Tzanakakis, E. S.
Fig.4.F

-
FC/FACS
-
Collected and cropped from PLoS One by CiteAb, provided under a CC-BY license
Image 1 of 7
In PLoS One on 12 December 2012 by Wu, J. & Tzanakakis, E. S.
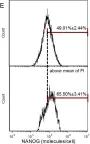
Fig.4.E

-
FC/FACS
-
Collected and cropped from PLoS One by CiteAb, provided under a CC-BY license
Image 1 of 7
In PLoS One on 12 December 2012 by Wu, J. & Tzanakakis, E. S.
Fig.5.B

-
FC/FACS
-
Collected and cropped from PLoS One by CiteAb, provided under a CC-BY license
Image 1 of 7
In PLoS One on 12 December 2012 by Wu, J. & Tzanakakis, E. S.
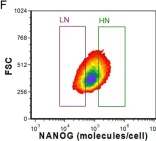
Fig.4.G

-
FC/FACS
-
Collected and cropped from PLoS One by CiteAb, provided under a CC-BY license
Image 1 of 7
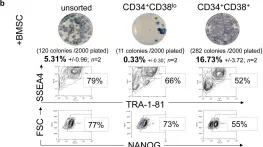
In PLoS One on 21 August 2012 by Park, T. S., Huo, J. S., et al.
Fig.2.B

-
FC/FACS
-
Collected and cropped from PLoS One by CiteAb, provided under a CC-BY license
Image 1 of 7