To produce abundant cell culture samples to generate large, standardized image datasets of human induced pluripotent stem (hiPS) cells, we developed an automated workflow on a Hamilton STAR liquid handler system. This was developed specifically for culturing hiPS cell lines expressing fluorescently tagged proteins, which we have used to study the principles by which cells establish and maintain robust dynamic localization of cellular structures. This protocol includes all details for the maintenance, passage and seeding of cells, as well as Matrigel coating of 6-well plastic plates and 96-well optical-grade, glass plates. We also developed an automated image-based hiPS cell colony segmentation and feature extraction pipeline to streamline the process of predicting cell count and selecting wells with consistent morphology for high-resolution three-dimensional (3D) microscopy. The imaging samples produced with this protocol have been used to study the integrated intracellular organization and cell-to-cell variability of hiPS cells to train and develop deep learning-based label-free predictions from transmitted-light microscopy images and to develop deep learning-based generative models of single-cell organization. This protocol requires some experience with robotic equipment. However, we provide details and source code to facilitate implementation by biologists less experienced with robotics. The protocol is completed in less than 10 h with minimal human interaction. Overall, automation of our cell culture procedures increased our imaging samples' standardization, reproducibility, scalability and consistency. It also reduced the need for stringent culturist training and eliminated culturist-to-culturist variability, both of which were previous pain points of our original manual pipeline workflow.
© 2023. Crown.
Product Citations: 31
In Nature Protocols on 1 February 2024 by Gregor, B. W., Coston, M. E., et al.
-
Stem Cells and Developmental Biology
In Human Cell on 1 November 2023 by Rani, S., Thamodaran, V., et al.
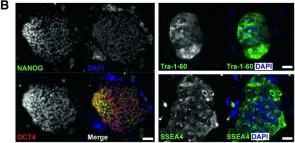
Diamond-Blackfan anemia (DBA) is a congenital hypoplastic anemia characterized by ineffective erythropoiesis. DBA is majorly caused by mutations in the ribosomal protein (RP) genes (Gadhiya and Wills in Diamond-Blackfan Anemia, https://www.statpearls.com/ ; 2023). A suitable disease model that yields a continuous supply of erythroid cells is required to study disease pathogenesis and drug discovery. Toward this, we reprogrammed dermal fibroblasts from a DBA patient with a heterozygous mutation c.22-23delAG in the RPS19 gene identified through exome sequencing. To generate induced pluripotent stem cells (iPSCs), we induced episomal expression of the reprogramming factors OTC3/4, L-MYC, LIN28, SOX2, and KLF4, and a p53 shRNA2. The DBA-iPSC line CSCRi006-A generated during this study was extensively characterized for its pluripotency and genome stability. The clone retained normal karyotype and showed high expression levels of pluripotency markers, OCT4, NANOG, SOX2, TRA-I-60, TRA-I-81, and SSEA4. It could differentiate into cells originating from all three germ cell layers, as identified by immunostaining for SOX17 (endoderm), Brachyury (mesoderm), and PAX6 (ectoderm). IPSCs provide a renewable source of cells for in vitro disease modeling. CSCRi006-A, a thoroughly characterized iPSC line carrying heterozygous RPS19 c.22-23delAG mutation, is a valuable cell line for the disease modeling of DBA. This iPSC line can be differentiated into different blood cell types to study the mechanisms of disease development and identify potential treatments.
© 2023. The Author(s) under exclusive licence to Japan Human Cell Society.
-
Homo sapiens (Human)
-
Stem Cells and Developmental Biology
In Nature Communications on 14 September 2023 by Mouti, M. A., Deng, S., et al.
Pancreatic cancer (PC), one of the most aggressive and life-threatening human malignancies, is known for its resistance to cytotoxic therapies. This is increasingly ascribed to the subpopulation of undifferentiated cells, known as pancreatic cancer stem cells (PCSCs), which display greater evolutionary fitness than other tumor cells to evade the cytotoxic effects of chemotherapy. PCSCs are crucial for tumor relapse as they possess 'stem cell-like' features that are characterized by self-renewal and differentiation. However, the molecular mechanisms that maintain the unique characteristics of PCSCs are poorly understood. Here, we identify the histone methyltransferase KMT2A as a physical binding partner of an RNA polymerase-associated PHF5A-PHF14-HMG20A-RAI1 protein subcomplex and an epigenetic regulator of PCSC properties and functions. Targeting the protein subcomplex in PCSCs with a KMT2A-WDR5 inhibitor attenuates their self-renewal capacity, cell viability, and in vivo tumorigenicity.
© 2023. Springer Nature Limited.
-
FC/FACS
-
Cancer Research
-
Stem Cells and Developmental Biology
SFPQ and Its Isoform as Potential Biomarker for Non-Small-Cell Lung Cancer.
In International Journal of Molecular Sciences on 6 August 2023 by Yang, L., Gilbertsen, A., et al.
Cancer markers are measurable molecules in the blood or tissue that are produced by tumor cells or immune cells in response to cancer progression. They play an important role in clinical diagnosis, prognosis, and anti-drug monitoring. Although DNA, RNA, and even physical images have been used, proteins continue to be the most common marker. There are currently no specific markers for lung cancer. Metastatic lung cancer, particularly non-small-cell lung cancer (NSCLC), is one of the most common causes of death. SFPQ, YY1, RTN4, RICTOR, LARP6, and HELLS are expressed at higher levels in cells from NSCLC than in control or cells from inflammatory diseases. SFPQ shows the most difference between the three cell types. Furthermore, the cytoplasmic isoform of SFPQ is only found in advanced cancers. We have developed ELISAs to detect SFPQ and the long and short isoforms. Evidence has shown that the short isoform exists primarily in cancers. Furthermore, immunocytometry studies and IHC analysis have revealed that SFPQ levels are consistent with ELISA results. In addition, enhanced DNA methylation in the SFPQ gene may facilitate the SFPQ expression differences between control and cancer cells. Considering this, elevated SFPQ level and the isoform location could serve as a cancer diagnostic and prognostic marker.
-
FC/FACS
-
Homo sapiens (Human)
-
Cancer Research
In Cell Death & Disease on 10 June 2023 by Domingo-Reinés, J., Montes, R., et al.
Pediatric Acute Myeloid Leukemia (AML) is a rare and heterogeneous disease characterized by a high prevalence of gene fusions as driver mutations. Despite the improvement of survival in the last years, about 50% of patients still experience a relapse. It is not possible to improve prognosis only with further intensification of chemotherapy, as come with a severe cost to the health of patients, often resulting in treatment-related death or long-term sequels. To design more effective and less toxic therapies we need a better understanding of pediatric AML biology. The NUP98-KDM5A chimeric protein is exclusively found in a particular subgroup of young pediatric AML patients with complex karyotypes and poor prognosis. In this study, we investigated the impact of NUP98-KDM5A expression on cellular processes in human Pluripotent Stem Cell models and a patient-derived cell line. We found that NUP98-KDM5A generates genomic instability through two complementary mechanisms that involve accumulation of DNA damage and direct interference of RAE1 activity during mitosis. Overall, our data support that NUP98-KDM5A promotes genomic instability and likely contributes to malignant transformation.
© 2023. The Author(s).
-
FC/FACS
-
Homo sapiens (Human)
-
Cancer Research
-
Cell Biology
In EMBO Rep on 5 November 2019 by Kim, J., Lana, B., et al.
Fig.1.B

-
ICC-IF
-
Homo sapiens (Human)
Collected and cropped from EMBO Rep by CiteAb, provided under a CC-BY license
Image 1 of 1