KCNQ1/Kv7, a low-voltage-gated K+ channel, regulates cardiac rhythm and glucose homeostasis. While KCNQ1 mutations are associated with long-QT syndrome and type2 diabetes, its function in human pancreatic cells remains controversial. We identified a homozygous KCNQ1 mutation (R397W) in an individual with permanent neonatal diabetes melitus (PNDM) without cardiovascular symptoms. To decipher the potential mechanism(s), we introduced the mutation into human embryonic stem cells and generated islet-like organoids (SC-islets) using CRISPR-mediated homology-repair. The mutation did not affect pancreatic differentiation, but affected channel function by increasing spike frequency and Ca2+ flux, leading to insulin hypersecretion. With prolonged culturing, the mutant islets decreased their secretion and gradually deteriorated, modeling a diabetic state, which accelerated by high glucose levels. The molecular basis was the downregulated expression of voltage-activated Ca2+ channels and oxidative phosphorylation. Our study provides a better understanding of the role of KCNQ1 in regulating insulin secretion and β-cell survival in hereditary diabetes pathology.© 2024 The Authors.
Product Citations: 11
In IScience on 19 July 2024 by Zhou, Z., Gong, M., et al.
Influence of feeder cells on transcriptomic analysis of pluripotent stem cells.
In Cell Proliferation on 1 February 2022 by Wan, H., Fu, R., et al.
Human pluripotent stem cells (hPSCs) are of great importance in both scientific research and regenerative medicine. The most classic and widely used culture method for hPSCs is co-culture with feeder cells, usually mouse embryonic fibroblasts. However, whether these feeder cell residues can affect the transcriptomic data analysis of hPSCs, especially gene or miRNA expression quantification, is still largely unknown.
In this study, reanalysis of published mRNA-Seq and miRNA-Seq data sets revealed the existence of feeder cell-derived reads in the hPSC transcriptomic samples. We identified potentially influenced human genes and miRNAs due to misalignment of sequencing fragments affected by mouse feeder cells. Furthermore, we developed an optimized miRNA analysis pipeline to avoid quantification bias from different miRNA isoforms in the same family. Finally, by comparing the levels of feeder cell residues in hPSC samples isolated by different methods, we found that fluorescence-activated cell sorting and adhesion methods were more effective in feeder cell removal than the gradient centrifugation method.
Collectively, our results demonstrate that feeder cell residues affect the transcriptomic data analysis of hPSCs. To minimize the impact of feeder cell contamination in hPSC samples, we provide solutions for both data analysis and sample preparation.
© 2022 The Authors. Cell Proliferation Published by John Wiley & Sons Ltd.
-
FC/FACS
-
Stem Cells and Developmental Biology
End-to-End Platform for Human Pluripotent Stem Cell Manufacturing.
In International Journal of Molecular Sciences on 21 December 2019 by Pandey, P. R., Tomney, A., et al.
Industrialization of stem-cell based therapies requires innovative solutions to close the gap between research and commercialization. Scalable cell production platforms are needed to reliably deliver the cell quantities needed during the various stages of development and commercial supply. Human pluripotent stem cells (hPSCs) are a key source material for generating therapeutic cell types. We have developed a closed, automated and scalable stirred tank bioreactor platform, capable of sustaining high fold expansion of hPSCs. Such a platform could facilitate the in-process monitoring and integration of online monitoring systems, leading to significantly reduced labor requirements and contamination risk. hPSCs are expanded in a controlled bioreactor using perfused xeno-free media. Cell harvest and concentration are performed in closed steps. The hPSCs can be cryopreserved to generate a bank of cells, or further processed as needed. Cryopreserved cells can be thawed into a two-dimensional (2D) tissue culture platform or a three-dimensional (3D) bioreactor to initiate a new expansion phase, or be differentiated to the clinically relevant cell type. The expanded hPSCs express hPSC-specific markers, have a normal karyotype and the ability to differentiate to the cells of the three germ layers. This end-to-end platform allows a large scale expansion of high quality hPSCs that can support the required cell demand for various clinical indications.
-
FC/FACS
-
Stem Cells and Developmental Biology
In Cell Reports on 9 April 2019 by Mair, B., Tomic, J., et al.
Human pluripotent stem cells (hPSCs) provide an invaluable tool for modeling diseases and hold promise for regenerative medicine. For understanding pluripotency and lineage differentiation mechanisms, a critical first step involves systematically cataloging essential genes (EGs) that are indispensable for hPSC fitness, defined as cell reproduction in this study. To map essential genetic determinants of hPSC fitness, we performed genome-scale loss-of-function screens in an inducible Cas9 H1 hPSC line cultured on feeder cells and laminin to identify EGs. Among these, we found FOXH1 and VENTX, genes that encode transcription factors previously implicated in stem cell biology, as well as an uncharacterized gene, C22orf43/DRICH1. hPSC EGs are substantially different from other human model cell lines, and EGs in hPSCs are highly context dependent with respect to different growth substrates. Our CRISPR screens establish parameters for genome-wide screens in hPSCs, which will facilitate the characterization of unappreciated genetic regulators of hPSC biology.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
-
Stem Cells and Developmental Biology
In Developmental Cell on 5 February 2018 by Böiers, C., Richardson, S. E., et al.
ETV6-RUNX1 is associated with childhood acute B-lymphoblastic leukemia (cALL) functioning as a first-hit mutation that initiates a clinically silent pre-leukemia in utero. Because lineage commitment hierarchies differ between embryo and adult, and the impact of oncogenes is cell-context dependent, we hypothesized that the childhood affiliation of ETV6-RUNX1 cALL reflects its origins in a progenitor unique to embryonic life. We characterize the first emerging B cells in first-trimester human embryos, identifying a developmentally restricted CD19-IL-7R+ progenitor compartment, which transitions from a myeloid to lymphoid program during ontogeny. This developmental series is recapitulated in differentiating human pluripotent stem cells (hPSCs), thereby providing a model for the initiation of cALL. Genome-engineered hPSCs expressing ETV6-RUNX1 from the endogenous ETV6 locus show expansion of the CD19-IL-7R+ compartment, show a partial block in B lineage commitment, and produce proB cells with aberrant myeloid gene expression signatures and potential: features (collectively) consistent with a pre-leukemic state.
Copyright © 2017 The Author(s). Published by Elsevier Inc. All rights reserved.
-
Cancer Research
-
Stem Cells and Developmental Biology
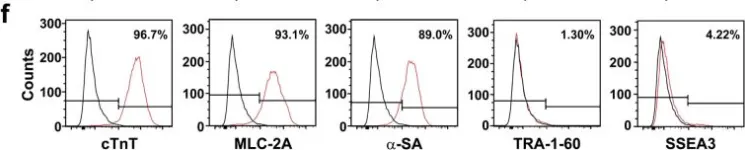
In Sci Rep on 23 October 2017 by Kim, H. S., Yoon, J. W., et al.
Fig.1.F

-
FC/FACS
-
Homo sapiens (Human)
Collected and cropped from Sci Rep by CiteAb, provided under a CC-BY license
Image 1 of 1