Hepatocytes are crucial for drug screening, disease modeling, and clinical transplantation, yet generating functional hepatocytes in vitro is challenging due to the difficulty of establishing their authentic gene regulatory networks (GRNs). We have previously developed a two-step lineage reprogramming strategy to generate functionally competent human induced hepatocytes (hiHeps), providing an effective model for studying the establishment of hepatocyte-specific GRNs. In this study, we utilized high-throughput single-cell RNA sequencing (scRNA-seq) to explore the cell-fate transition and the establishment of hepatocyte-specific GRNs involved in the two-step reprogramming process. Our findings revealed that the late stage of the reprogramming process mimics the natural trajectory of liver development, exhibiting similar transcriptional waves of developmental genes. CD24 and DLK1 were identified as surface markers enriching two distinct hepatic progenitor populations respectively. Lipid metabolism emerged as a key enhancer of hiHeps maturation. Furthermore, transcription factors HNF4A and HHEX were identified as pivotal gatekeepers directing cell fate decisions between hepatocytes and intestinal cells. Collectively, this study provides valuable insights into the establishment of hepatocyte-specific GRNs during hiHeps induction at single-cell resolution, facilitating more efficient production of functional hepatocytes for therapeutic applications.
© 2025. The Author(s).
Product Citations: 24
In Cellular and Molecular Life Sciences : CMLS on 6 April 2025 by Jiang, N., Li, G., et al.
-
Biochemistry and Molecular biology
-
Stem Cells and Developmental Biology
In Nature Communications on 17 June 2024 by Razavipour, S. F., Yoon, H., et al.
In many cancers, a stem-like cell subpopulation mediates tumor initiation, dissemination and drug resistance. Here, we report that cancer stem cell (CSC) abundance is transcriptionally regulated by C-terminally phosphorylated p27 (p27pT157pT198). Mechanistically, this arises through p27 co-recruitment with STAT3/CBP to gene regulators of CSC self-renewal including MYC, the Notch ligand JAG1, and ANGPTL4. p27pTpT/STAT3 also recruits a SIN3A/HDAC1 complex to co-repress the Pyk2 inhibitor, PTPN12. Pyk2, in turn, activates STAT3, creating a feed-forward loop increasing stem-like properties in vitro and tumor-initiating stem cells in vivo. The p27-activated gene profile is over-represented in STAT3 activated human breast cancers. Furthermore, mammary transgenic expression of phosphomimetic, cyclin-CDK-binding defective p27 (p27CK-DD) increases mammary duct branching morphogenesis, yielding hyperplasia and microinvasive cancers that can metastasize to liver, further supporting a role for p27pTpT in CSC expansion. Thus, p27pTpT interacts with STAT3, driving transcriptional programs governing stem cell expansion or maintenance in normal and cancer tissues.
© 2024. The Author(s).
-
FC/FACS
-
Cancer Research
-
Stem Cells and Developmental Biology
In Frontiers in Oncology on 19 December 2023 by Knutsen, E., Das Sajib, S., et al.
Epithelial-mesenchymal transition (EMT) is a cellular plasticity program critical for embryonic development and tissue regeneration, and aberrant EMT is associated with disease including cancer. The high degree of plasticity in the mammary epithelium is reflected in extensive heterogeneity among breast cancers. Here, we have analyzed RNA-sequencing data from three different mammary epithelial cell line-derived EMT models and identified a robust mammary EMT gene expression signature that separates breast cancers into distinct subgroups. Most strikingly, the basal-like breast cancers form two subgroups displaying partial-EMT and post-EMT gene expression patterns. We present evidence that key EMT-associated transcription factors play distinct roles at different stages of EMT in mammary epithelial cells.
Copyright © 2023 Knutsen, Das Sajib, Fiskaa, Lorens, Gudjonsson, Mælandsmo, Johansen, Seternes and Perander.
-
FC/FACS
-
Homo sapiens (Human)
In Cancers on 12 April 2023 by Ben-Yaakov, H., Meshel, T., et al.
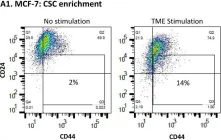
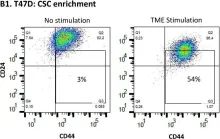
Hormone receptor-positive and HER2-negative (HR+/HER2-; luminal A) tumors are prevalent in breast cancer. Our past studies demonstrated that "TME Stimulation" (estrogen + TNFα + EGF, representing three arms of the tumor microenvironment, TME) has enriched metastasis-forming cancer stem cells (CSCs) in HR+/HER2- human breast cancer cells. Here, following information obtained by RNAseq analyses of TME-stimulated CSCs and Non-CSCs, we found that TME Stimulation has induced the activation of S727-STAT3, Y705-STAT3, STAT1 and p65. Upon TME Stimulation, stattic (STAT3 inhibitor) usage demonstrated that Y705-STAT3 activation negatively controlled CSC enrichment and epithelial-to-mesenchymal transition (EMT) traits, while inducing CXCL8 (IL-8) and PD-L1 expression. However, STAT3 knock-down (siSTAT3) had no effect on these functions; in terms of CSC enrichment, p65 had down-regulatory roles that compensated for the loss of an entire STAT3 protein. Y705-STAT3 and p65 acted additively in reducing CSC enrichment, and Y705A-STAT3 variant + sip65 has enriched chemo-resistant CSCs. Clinical data analyses revealed an inverse correlation between Y705-STAT3 + p65 phosphorylation and CSC signature in luminal A patients, and connection to improved disease course. Overall, we find regulatory roles for Y705-STAT3 and p65 in TME-stimulated HR+/HER2- tumors, with the ability to limit CSC enrichment. These findings raise concerns about using inhibitors of STAT3 and p65 as therapeutic strategies in the clinic.
-
FC/FACS
-
Homo sapiens (Human)
-
Cancer Research
-
Stem Cells and Developmental Biology
In Journal of Translational Medicine on 1 April 2023 by Zhao, H., Jiang, R., et al.
Extracellular vesicles (EVs) transport biologically active molecules, and represent a recently identified way of intercellular communication. Recent evidence has also reported that EVs shed by cancer stem cells (CSCs) make a significant contribution to carcinogenesis and metastasis. Here, this study aims to explore the possible molecular mechanism of CSCs-EVs in gastric cancer (GC) by mediating intratumor communication network.
CSCs and non-stem cancer cells (NSCCs) were sorted from GC cells, and EVs were isolated from CSCs. H19 was knocked down in CSCs, and CSCs-EVs or CSCs-EVs containing shRNA-H19 (CSCs-EVs-sh-H19) were co-cultured with NSCCs, followed by evaluation of the malignant behaviors and stemness of NSCCs. Mouse models of GC were established and injected with CSCs-EVs from sh-H19-treated NSCCs in vivo.
CSCs had notable self-renewal and tumorigenic capacity compared with NSCCs. CSCs promoted the malignant behaviors of NSCCs and expression of stemness marker proteins through secretion of EVs. Inhibited secretion of CSCs-EVs curtailed the tumorigenicity and metastasis of NSCCs in vivo. H19 could be delivered by CSCs-EVs into NSCCs. H19 promoted the malignant behaviors of NSCCs and stemness marker protein expression in vitro along with tumorigenicity and liver metastasis in vivo, which was mechanistically associated with activation of the YAP/CDX2 signaling axis.
Taken together, the present study points to the importance of a novel regulatory axis H19/YAP/CDX2 in carcinogenic and metastatic potential of CSCs-EVs in GC, which may be potential targets for anticancer therapy.
© 2023. The Author(s).
-
FC/FACS
-
Cancer Research
-
Stem Cells and Developmental Biology
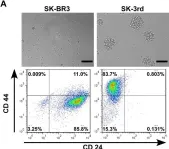
In Cancers (Basel) on 12 April 2023 by Ben-Yaakov, H., Meshel, T., et al.
Fig.1.A

-
FC/FACS
-
Homo sapiens (Human)
Collected and cropped from Cancers (Basel) by CiteAb, provided under a CC-BY license
Image 1 of 5
In Cancers (Basel) on 12 April 2023 by Ben-Yaakov, H., Meshel, T., et al.
Fig.1.B

-
FC/FACS
-
Homo sapiens (Human)
Collected and cropped from Cancers (Basel) by CiteAb, provided under a CC-BY license
Image 1 of 5
In Elife on 19 June 2019 by Essex, A., Pineda, J., et al.
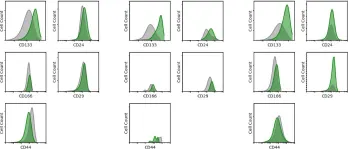
Fig.1.C

-
FC/FACS
-
Homo sapiens (Human)
Collected and cropped from Elife by CiteAb, provided under a CC-BY license
Image 1 of 5
In Elife on 19 June 2019 by Essex, A., Pineda, J., et al.
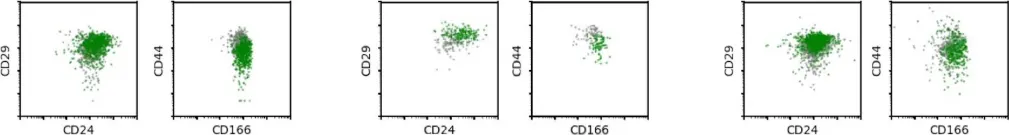
Fig.1.D

-
FC/FACS
-
Homo sapiens (Human)
Collected and cropped from Elife by CiteAb, provided under a CC-BY license
Image 1 of 5
In PLoS One on 9 June 2012 by Li, J., Zucker, S., et al.
Fig.1.A

-
FC/FACS
-
Collected and cropped from PLoS One by CiteAb, provided under a CC-BY license
Image 1 of 5