A combination of hematopoietic stem cell (HSC)- and endothelial cell (EC)-lineage-tracing mouse models enables us to determine blood cell origins. We present a protocol to induce cell labeling in vivo and to trace labeled hematopoietic cells to segregate their origins. We describe the steps for harvesting various hematopoietic tissues, antibody staining, and analyzing the Tomato+ percentages within each immune cell population. We also show how to estimate HSC- and EC-derived percentages of the target cell populations. For complete details on the use and execution of this protocol, please refer to Kobayashi et al.1.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Product Citations: 21
In STAR Protocols on 20 December 2024 by Syed, A., Kobayashi, M., et al.
In Developmental Cell on 6 May 2024 by Fowler, J. L., Zheng, S. L., et al.
The developmental origin of blood-forming hematopoietic stem cells (HSCs) is a longstanding question. Here, our non-invasive genetic lineage tracing in mouse embryos pinpoints that artery endothelial cells generate HSCs. Arteries are transiently competent to generate HSCs for 2.5 days (∼E8.5-E11) but subsequently cease, delimiting a narrow time frame for HSC formation in vivo. Guided by the arterial origins of blood, we efficiently and rapidly differentiate human pluripotent stem cells (hPSCs) into posterior primitive streak, lateral mesoderm, artery endothelium, hemogenic endothelium, and >90% pure hematopoietic progenitors within 10 days. hPSC-derived hematopoietic progenitors generate T, B, NK, erythroid, and myeloid cells in vitro and, critically, express hallmark HSC transcription factors HLF and HOXA5-HOXA10, which were previously challenging to upregulate. We differentiated hPSCs into highly enriched HLF+ HOXA+ hematopoietic progenitors with near-stoichiometric efficiency by blocking formation of unwanted lineages at each differentiation step. hPSC-derived HLF+ HOXA+ hematopoietic progenitors could avail both basic research and cellular therapies.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
-
FC/FACS
-
Homo sapiens (Human)
-
Stem Cells and Developmental Biology
In Arteriosclerosis, Thrombosis, and Vascular Biology on 1 March 2024 by Mao, A., Zhang, K., et al.
Single-cell RNA-Seq analysis can determine the heterogeneity of cells between different tissues at a single-cell level. Coronary artery endothelial cells (ECs) are important to coronary blood flow. However, little is known about the heterogeneity of coronary artery ECs, and cellular identity responses to flow. Identifying endothelial subpopulations will contribute to the precise localization of vascular endothelial subpopulations, thus enabling the precision of vascular injury treatment.
Here, we performed a single-cell RNA sequencing of 31 962 cells and functional assays of 3 branches of the coronary arteries (right coronary artery/circumflex left coronary artery/anterior descending left coronary artery) in wild-type mice.
We found a compendium of 7 distinct cell types in mouse coronary arteries, mainly ECs, granulocytes, cardiac myocytes, smooth muscle cells, lymphocytes, myeloid cells, and fibroblast cells, and showed spatial heterogeneity between arterial branches. Furthermore, we revealed a subpopulation of coronary artery ECs, CD133+TRPV4high ECs. TRPV4 (transient receptor potential vanilloid 4) in CD133+TRPV4high ECs is important for regulating vasodilation and coronary blood flow.
Our study elucidates the nature and range of coronary arterial cell diversity and highlights the importance of coronary CD133+TRPV4high ECs in regulating coronary vascular tone.
-
Mus musculus (House mouse)
-
Cardiovascular biology
-
Genetics
In Research (Washington, D.C.) on 3 July 2023 by Liu, Q., Long, Q., et al.
Adipose browning has demonstrated therapeutic potentials in several diseases. Here, by conducting transcriptomic profiling at the single-cell and single-nucleus resolution, we reconstituted the cellular atlas in mouse inguinal subcutaneous white adipose tissue (iWAT) at thermoneutrality or chronic cold condition. All major nonimmune cells within the iWAT, including adipose stem and progenitor cells (ASPCs), mature adipocytes, endothelial cells, Schwann cells, and smooth muscle cells, were recovered, allowing us to uncover an overall and detailed blueprint for transcriptomes and intercellular cross-talks and the dynamics during white adipose tissue brown remodeling. Our findings also unravel the existence of subpopulations in mature adipocytes, ASPCs, and endothelial cells, as well as new insights on their interconversion and reprogramming in response to cold. The adipocyte subpopulation competent of major histocompatibility complex class II (MHCII) antigen presentation is potentiated. Furthermore, a subcluster of ASPC with CD74 expression was identified as the precursor of this MHCII+ adipocyte. Beige adipocytes are transdifferented from preexisting lipid generating adipocytes, which exhibit developmental trajectory from de novo differentiation of amphiregulin cells (Aregs). Two distinct immune-like endothelial subpopulations are present in iWAT and are responsive to cold. Our data reveal fundamental changes during cold-evoked adipose browning.
Copyright © 2023 Qing Liu et al.
-
FC/FACS
-
Mus musculus (House mouse)
-
Genetics
Directed differentiation of mouse pluripotent stem cells into functional lung-specific mesenchyme.
In Nature Communications on 13 June 2023 by Alber, A. B., Marquez, H. A., et al.
While the generation of many lineages from pluripotent stem cells has resulted in basic discoveries and clinical trials, the derivation of tissue-specific mesenchyme via directed differentiation has markedly lagged. The derivation of lung-specific mesenchyme is particularly important since this tissue plays crucial roles in lung development and disease. Here we generate a mouse induced pluripotent stem cell (iPSC) line carrying a lung-specific mesenchymal reporter/lineage tracer. We identify the pathways (RA and Shh) necessary to specify lung mesenchyme and find that mouse iPSC-derived lung mesenchyme (iLM) expresses key molecular and functional features of primary developing lung mesenchyme. iLM recombined with engineered lung epithelial progenitors self-organizes into 3D organoids with juxtaposed layers of epithelium and mesenchyme. Co-culture increases yield of lung epithelial progenitors and impacts epithelial and mesenchymal differentiation programs, suggesting functional crosstalk. Our iPSC-derived population thus provides an inexhaustible source of cells for studying lung development, modeling diseases, and developing therapeutics.
© 2023. The Author(s).
-
Mus musculus (House mouse)
-
Stem Cells and Developmental Biology
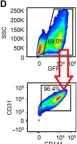
In Sci Adv on 3 February 2023 by Fan, M., Yang, K., et al.
Fig.2.D

-
FC/FACS
-
Collected and cropped from Sci Adv by CiteAb, provided under a CC-BY license
Image 1 of 1