Hidradenitis suppurativa (HS) is a relatively common and highly morbid inflammatory skin disease. Due to the relatively limited understanding of HS's pathogenesis, there are currently insufficient treatment options available, and many patients' medical needs are not being met. This is partly due to the historical scarcity of ex vivo assays and animal models that accurately recapitulate the disease. Thus, we have developed a standardised whole-tissue explant model of HS to examine its pathogenic mechanisms and the efficacy of potential treatments within intact human tissue. We measured cytokine protein and RNA within whole tissue maintained in an agar-media solution, finding that IL-6 and IL-8 concentrations trended upwards in both HS explants and healthy controls, while IL-17A, IL-1β, and TNF-α exhibited increases in HS tissue alone. We also show that the explants were responsive to treatment with both dexamethasone and IL-2. Not only do our results show that this model effectively delivers treatments throughout the explants, but they also elucidate which cytokines are related to the explant process regardless of tissue state and which are related to HS tissue specifically, laying the groundwork for future implementations of this model.
© 2025 The Author(s). Experimental Dermatology published by John Wiley & Sons Ltd.
Product Citations: 38
The Inflammatory Landscape of a Whole-Tissue Explant Model of Hidradenitis Suppurativa.
In Experimental Dermatology on 1 February 2025 by Leboit, P. E., Patel, D. U., et al.
-
Immunology and Microbiology
In Cells on 26 October 2024 by Ahmady, F., Curpen, P., et al.
Inhibitory receptors are critical for regulating immune cell function. In cancer, these receptors are often over-expressed on the cell surface of T and NK cells, leading to reduced anti-tumor activity. Here, through the analysis of 11 commonly studied checkpoint and inhibitory receptors, we discern that only HAVCR2 (TIM3) and ENTPD1 (CD39) display significantly greater gene expression in glioblastoma compared to normal brain and lower grade glioma. Cell surface TIM-3, but not ENTPD1, was also elevated on activated CD4+ and CD8+ T cells, as well as on NK cells from glioblastoma patients compared to healthy donor T and NK cells. A subsequent analysis of molecules known to co-ordinate TIM-3 function and regulation was performed, which revealed that BAT3 expression was significantly reduced in CD4+ and CD8+ T cells, as well as NK cells from glioblastoma patients compared to counterparts from healthy donors. These pro-inhibitory changes are also correlated with reduced levels of the activation marker CD69 and the pro-inflammatory cytokine IFNγ in CD4+ and CD8+ T cells, as well as NK cells from glioblastoma patients. Collectively, these data reveal that glioblastoma-mediated CD4+ and CD8+ T cell and NK cell suppression is due, at least in part, to dysregulated TIM-3 and BAT3 expression and the associated downstream immunoregulatory and dysfunctional effects.
-
Cell Biology
Protocol to construct humanized mice with adult CD34+ hematopoietic stem and progenitor cells.
In STAR Protocols on 20 September 2024 by Yu, C. I., Maser, R., et al.
Humanized mice, defined as mice with human immune systems, have become an emerging model to study human hematopoiesis, infectious disease, and cancer. Here, we describe the techniques to generate humanized NSGF6 mice using adult human CD34+ hematopoietic stem and progenitor cells (HSPCs). We describe steps for constructing and monitoring the engraftment of humanized mice. We then detail procedures for tissue processing and immunophenotyping by flow cytometry to evaluate the multilineage hematopoietic differentiation. For complete details on the use and execution of this protocol, please refer to Yu et al.1.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Preprint on BioRxiv : the Preprint Server for Biology on 1 July 2024 by Croce, G., Lani, R., et al.
T cells targeting epitopes in infectious diseases or cancer play a central role in spontaneous and therapy-induced immune responses. T-cell epitope recognition is mediated by the binding of the T-Cell Receptor (TCR) and TCRs recognizing clinically relevant epitopes are promising for T-cell based therapies. Starting from one of the few known TCRs targeting the cancer-testis antigen NY-ESO-1 157-165 epitope, we built large phage display libraries of TCRs with randomized Complementary Determining Region 3 of the β chain. The TCR libraries were panned against the NY-ESO-1 epitope, which enabled us to collect thousands of epitope-specific TCR sequences. We then trained a machine learning TCR-epitope interaction predictor with this data and could identify several epitope-specific TCRs directly from TCR repertoires. Cellular binding and functional assays revealed that the predicted TCRs displayed activity towards the NY-ESO-1 epitope and no detectable cross-reactivity with self-peptides.
PGE2 inhibits TIL expansion by disrupting IL-2 signalling and mitochondrial function.
In Nature on 1 May 2024 by Morotti, M., Grimm, A. J., et al.
Expansion of antigen-experienced CD8+ T cells is critical for the success of tumour-infiltrating lymphocyte (TIL)-adoptive cell therapy (ACT) in patients with cancer1. Interleukin-2 (IL-2) acts as a key regulator of CD8+ cytotoxic T lymphocyte functions by promoting expansion and cytotoxic capability2,3. Therefore, it is essential to comprehend mechanistic barriers to IL-2 sensing in the tumour microenvironment to implement strategies to reinvigorate IL-2 responsiveness and T cell antitumour responses. Here we report that prostaglandin E2 (PGE2), a known negative regulator of immune response in the tumour microenvironment4,5, is present at high concentrations in tumour tissue from patients and leads to impaired IL-2 sensing in human CD8+ TILs via the PGE2 receptors EP2 and EP4. Mechanistically, PGE2 inhibits IL-2 sensing in TILs by downregulating the IL-2Rγc chain, resulting in defective assembly of IL-2Rβ-IL2Rγc membrane dimers. This results in impaired IL-2-mTOR adaptation and PGC1α transcriptional repression, causing oxidative stress and ferroptotic cell death in tumour-reactive TILs. Inhibition of PGE2 signalling to EP2 and EP4 during TIL expansion for ACT resulted in increased IL-2 sensing, leading to enhanced proliferation of tumour-reactive TILs and enhanced tumour control once the cells were transferred in vivo. Our study reveals fundamental features that underlie impairment of human TILs mediated by PGE2 in the tumour microenvironment. These findings have therapeutic implications for cancer immunotherapy and cell therapy, and enable the development of targeted strategies to enhance IL-2 sensing and amplify the IL-2 response in TILs, thereby promoting the expansion of effector T cells with enhanced therapeutic potential.
© 2024. The Author(s).
-
FC/FACS
-
Homo sapiens (Human)
-
Cell Biology
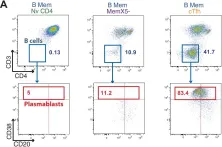
In MBio on 8 May 2018 by Claireaux, M., Galperin, M., et al.
Fig.4.A

-
FC/FACS
-
Homo sapiens (Human)
Collected and cropped from MBio by CiteAb, provided under a CC-BY license
Image 1 of 1