High levels of the polyunsaturated fatty acid arachidonic acid (AA) within the ovarian carcinoma (OC) microenvironment correlate with reduced relapse-free survival. Furthermore, OC progression is tied to compromised immunosurveillance, partially attributed to the impairment of natural killer (NK) cells. However, potential connections between AA and NK cell dysfunction in OC have not been studied.
We employed a combination of phosphoproteomics, transcriptional profiling and biological assays to investigate AA's impact on NK cell functions.
AA (i) disrupts interleukin-2/15-mediated expression of pro-inflammatory genes by inhibiting STAT1-dependent signaling, (ii) hampers signaling by cytotoxicity receptors through disruption of their surface expression, (iii) diminishes phosphorylation of NKG2D-induced protein kinases, including ERK1/2, LYN, MSK1/2 and STAT1, and (iv) alters reactive oxygen species production by transcriptionally upregulating detoxification. These modifications lead to a cessation of NK cell proliferation and a reduction in cytotoxicity.
Our findings highlight significant AA-induced alterations in the signaling network that regulates NK cell activity. As low expression of several NK cell receptors correlates with shorter OC patient survival, these findings suggest a functional linkage between AA, NK cell dysfunction and OC progression.
© 2024. The Author(s).
Product Citations: 8
In Cell Communication and Signaling : CCS on 19 November 2024 by Hammoud, M. K., Meena, C., et al.
-
FC/FACS
-
Homo sapiens (Human)
-
Endocrinology and Physiology
In Cell on 1 February 2024 by Li, Z., Ma, R., et al.
The therapeutic potential for human type 2 innate lymphoid cells (ILC2s) has been underexplored. Although not observed in mouse ILC2s, we found that human ILC2s secrete granzyme B (GZMB) and directly lyse tumor cells by inducing pyroptosis and/or apoptosis, which is governed by a DNAM-1-CD112/CD155 interaction that inactivates the negative regulator FOXO1. Over time, the high surface density expression of CD155 in acute myeloid leukemia cells impairs the expression of DNAM-1 and GZMB, thus allowing for immune evasion. We describe a reliable platform capable of up to 2,000-fold expansion of human ILC2s within 4 weeks, whose molecular and cellular ILC2 profiles were validated by single-cell RNA sequencing. In both leukemia and solid tumor models, exogenously administered expanded human ILC2s show significant antitumor effects in vivo. Collectively, we demonstrate previously unreported properties of human ILC2s and identify this innate immune cell subset as a member of the cytolytic immune effector cell family.
Copyright © 2023 Elsevier Inc. All rights reserved.
-
Cancer Research
Secretomes of M1 and M2 macrophages decrease the release of neutrophil extracellular traps.
In Scientific Reports on 20 September 2023 by Manda-Handzlik, A., Cieloch, A., et al.
The release of neutrophil extracellular traps (NETs) can be either beneficial or detrimental for the host, thus it is necessary to maintain a balance between formation and clearance of NETs. Multiple physiological factors eliciting NET release have been identified, yet the studies on natural signals limiting NET formation have been scarce. Accordingly, our aim was to analyze whether cytokines or immune cells can inhibit NET formation. To that end, human granulocytes were incubated with interleukin (IL)-4, IL-10, transforming growth factor beta-2 or adenosine and then stimulated to release NETs. Additionally, neutrophils were cultured in the presence of natural killer (NK) cells, regulatory T cells (Tregs), pro-inflammatory or anti-inflammatory macrophages (M1 or M2 macrophages), or in the presence of NK/Tregs/M1 macrophages or M2 macrophages-conditioned medium and subsequently stimulated to release NETs. Our studies showed that secretome of M1 and M2 macrophages, but not of NK cells and Tregs, diminishes NET formation. Co-culture experiments did not reveal any effect of immune cells on NET release. No effect of cytokines or adenosine on NET release was found. This study highlights the importance of paracrine signaling at the site of infection and is the first to show that macrophage secretome can regulate NET formation.
© 2023. Springer Nature Limited.
Untimely TGFβ responses in COVID-19 limit antiviral functions of NK cells.
In Nature on 1 December 2021 by Witkowski, M., Tizian, C., et al.
SARS-CoV-2 is a single-stranded RNA virus that causes COVID-19. Given its acute and often self-limiting course, it is likely that components of the innate immune system play a central part in controlling virus replication and determining clinical outcome. Natural killer (NK) cells are innate lymphocytes with notable activity against a broad range of viruses, including RNA viruses1,2. NK cell function may be altered during COVID-19 despite increased representation of NK cells with an activated and adaptive phenotype3,4. Here we show that a decline in viral load in COVID-19 correlates with NK cell status and that NK cells can control SARS-CoV-2 replication by recognizing infected target cells. In severe COVID-19, NK cells show defects in virus control, cytokine production and cell-mediated cytotoxicity despite high expression of cytotoxic effector molecules. Single-cell RNA sequencing of NK cells over the time course of the COVID-19 disease spectrum reveals a distinct gene expression signature. Transcriptional networks of interferon-driven NK cell activation are superimposed by a dominant transforming growth factor-β (TGFβ) response signature, with reduced expression of genes related to cell-cell adhesion, granule exocytosis and cell-mediated cytotoxicity. In severe COVID-19, serum levels of TGFβ peak during the first two weeks of infection, and serum obtained from these patients severely inhibits NK cell function in a TGFβ-dependent manner. Our data reveal that an untimely production of TGFβ is a hallmark of severe COVID-19 and may inhibit NK cell function and early control of the virus.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
-
COVID-19
In PLoS ONE on 23 October 2019 by Nakid-Cordero, C., Arzouk, N., et al.
Kidney transplant recipients (KTRs) abnormally replicate the Epstein Barr Virus (EBV). To better understand how long-term immunosuppression impacts the immune control of this EBV re-emergence, we systematically compared 10 clinically stable KTRs to 30 healthy controls (HCs). The EBV-specific T cell responses were determined in both groups by multiparameter flow cytometry with intra cellular cytokine staining (KTRs n = 10; HCs n = 15) and ELISpot-IFNγ assays (KTRs n = 7; HCs n = 7). The T/B/NK cell counts (KTRs n = 10; HCs n = 30) and the NK/T cell differentiation and activation phenotypes (KTRs n = 10; HCs n = 15/30) were also measured. We show that in KTRs, the Th1 effector CD4+ T cell responses against latent EBV proteins are weak (2/7 responders). Conversely, the frequencies total EBV-specific CD8+T cells are conserved in KTRs (n = 10) and span a wider range of EBNA-3A peptides (5/7responders) than in HCs (5/7responders). Those modifications of the EBV-specific T cell response were associated with a profound CD4+ T cell lymphopenia in KTRs compared to HCs, involving the naïve CD4+ T cell subset, and a persistent activation of highly-differentiated senescent CD8+ T cells. The proportion of total NK / CD8+ T cells expressing PD-1 was also increased in KTRs. Noteworthy, PD-1 expression on CD8+ T cells normalized with time after transplantation. In conclusion, we show modifications of the EBV-specific cellular immunity in long term transplant recipients. This may be the result of both persistent EBV antigenic stimulation and profound immunosuppression induced by anti-rejection treatments. These findings provide new insights into the immunopathology of EBV infection after renal transplantation.
-
Homo sapiens (Human)
-
Immunology and Microbiology
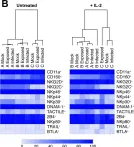
In PLoS Pathog on 1 June 2019 by Campbell, T. M., McSharry, B. P., et al.
Fig.4.B

-
WB
-
Collected and cropped from PLoS Pathog by CiteAb, provided under a CC-BY license
Image 1 of 1