Variant histone H3.3 is thought to be critical for survival of many cells, since it is deposited in expressed genes, a feature different from core histones. For example, H3.3 deletion leads to embryonic lethality in mice. However, requirement of H3.3 in later stage of development has remained unclear. The aim of this work was to elucidate the role of H3.3 for development of myeloid lineage, important for innate immunity. We conditionally knocked out (cKO) the H3.3 genes in myeloid progenitor cells differentiating into bone marrow derived macrophages (BMDMs). Progenitor cells lacking H3.3 were defective in replication, suffered from extensive DNA damage, and underwent apoptosis. Surviving H3.3 cKO cells expressed many interferon stimulated genes (ISGs) throughout differentiation. Further, H3.3 cKO BMDMs possessed chromatin accessible sites, and histone posttranslational modifications consistent with the gene expression profiles, Accordingly, H3.3 cKO BMDMs retained general nucleosomal structure genome wide. In summary, H3.3 is required for proliferation of myeloid progenitor cells, but is in large part dispensable for differentiation of BMDMs.
Product Citations: 29
Histone H3.3 ensures cell proliferation and genomic stability during myeloid cell development
Preprint on BioRxiv : the Preprint Server for Biology on 26 December 2024 by Chauhan, S., Dey, A., et al.
-
Genetics
High-resolution kinetic characterization of the RIG-I-signaling pathway and the antiviral response.
In Life Science Alliance on 1 October 2023 by Burkart, S. S., Schweinoch, D., et al.
RIG-I recognizes viral dsRNA and activates a cell-autonomous antiviral response. Upon stimulation, it triggers a signaling cascade leading to the production of type I and III IFNs. IFNs are secreted and signal to elicit the expression of IFN-stimulated genes, establishing an antiviral state of the cell. The topology of this pathway has been studied intensively, however, its exact dynamics are less understood. Here, we employed electroporation to synchronously activate RIG-I, enabling us to characterize cell-intrinsic innate immune signaling at a high temporal resolution. Employing IFNAR1/IFNLR-deficient cells, we could differentiate primary RIG-I signaling from secondary signaling downstream of the IFN receptors. Based on these data, we developed a comprehensive mathematical model capable of simulating signaling downstream of dsRNA recognition by RIG-I and the feedback and signal amplification by IFN. We further investigated the impact of viral antagonists on signaling dynamics. Our work provides a comprehensive insight into the signaling events that occur early upon virus infection and opens new avenues to study and disentangle the complexity of the host-virus interface.
© 2023 Burkart et al.
-
Homo sapiens (Human)
Human IRF1 governs macrophagic IFN-γ immunity to mycobacteria.
In Cell on 2 February 2023 by Rosain, J., Neehus, A. L., et al.
Inborn errors of human IFN-γ-dependent macrophagic immunity underlie mycobacterial diseases, whereas inborn errors of IFN-α/β-dependent intrinsic immunity underlie viral diseases. Both types of IFNs induce the transcription factor IRF1. We describe unrelated children with inherited complete IRF1 deficiency and early-onset, multiple, life-threatening diseases caused by weakly virulent mycobacteria and related intramacrophagic pathogens. These children have no history of severe viral disease, despite exposure to many viruses, including SARS-CoV-2, which is life-threatening in individuals with impaired IFN-α/β immunity. In leukocytes or fibroblasts stimulated in vitro, IRF1-dependent responses to IFN-γ are, both quantitatively and qualitatively, much stronger than those to IFN-α/β. Moreover, IRF1-deficient mononuclear phagocytes do not control mycobacteria and related pathogens normally when stimulated with IFN-γ. By contrast, IFN-α/β-dependent intrinsic immunity to nine viruses, including SARS-CoV-2, is almost normal in IRF1-deficient fibroblasts. Human IRF1 is essential for IFN-γ-dependent macrophagic immunity to mycobacteria, but largely redundant for IFN-α/β-dependent antiviral immunity.
Crown Copyright © 2022. Published by Elsevier Inc. All rights reserved.
-
WB
-
Homo sapiens (Human)
-
Immunology and Microbiology
Preprint on BioRxiv : the Preprint Server for Biology on 5 August 2022 by Burkart, S. S., Schweinoch, D., et al.
h4>ABSTRACT/h4> The pattern recognition receptor RIG-I is essential for the recognition of viral dsRNA and the activation of a cell-autonomous antiviral response. Upon stimulation, RIG-I triggers a signaling cascade leading to the expression of cytokines, most prominently type I and III interferons (IFNs). IFNs are secreted and signal in an auto- and paracrine manner to trigger the expression of a large variety of IFN-stimulated genes, which in concert establish an antiviral state of the cell. While the topology of this pathway has been studied quite intensively, the dynamics, particularly of the RIG-I-mediated IFN induction, is much less understood. Here, we employed electroporation-based transfection to synchronously activate the RIG-I signaling pathway, enabling us to characterize the kinetics and dynamics of cell-intrinsic innate immune signaling to virus infections. By employing an A549 IFNAR1/IFNLR deficient cell line, we could analyze the difference between the primary RIG-I signaling phase and the secondary signaling phase downstream of the IFN receptors. We further used our quantitative data to set up and calibrate a comprehensive dynamic mathematical model of the RIG-I and IFN signaling pathways. This model accurately predicts the kinetics of signaling events downstream of dsRNA recognition by RIG-I as well as the feedback and signal amplification by secreted IFN and JAK/STAT signaling. We have furthermore investigated the impact of various viral immune antagonists on the signaling dynamics experimentally, and we utilized the here described modelling approach to simulate and in silico study these critical virus-host interactions. Our work provides a comprehensive insight into the signaling events occurring early upon virus infection and opens up new avenues to study and disentangle the complexity of the host-virus interface.
-
WB
-
Homo sapiens (Human)
In International Journal of Molecular Sciences on 21 June 2022 by Jungwirth, J., Häring, C., et al.
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causing the coronavirus disease-19 (COVID-19) is still challenging healthcare systems and societies worldwide. While vaccines are available, therapeutic strategies are developing and need to be adapted to each patient. Many clinical approaches focus on the repurposing of approved therapeutics against other diseases. However, the efficacy of these compounds on viral infection or even harmful secondary effects in the context of SARS-CoV-2 infection are sparsely investigated. Similarly, adverse effects of commonly used therapeutics against lifestyle diseases have not been studied in detail. Using mono cell culture systems and a more complex chip model, we investigated the effects of the acetylsalicylic acid (ASA) salt D,L-lysine-acetylsalicylate + glycine (LASAG) on SARS-CoV-2 infection in vitro. ASA is commonly known as Aspirin® and is one of the most frequently used medications worldwide. Our data indicate an inhibitory effect of LASAG on SARS-CoV-2 replication and SARS-CoV-2-induced expression of pro-inflammatory cytokines and coagulation factors. Remarkably, our data point to an additive effect of the combination of LASAG and the antiviral acting drug remdesivir on SARS-CoV-2 replication in vitro.
-
WB
-
Homo sapiens (Human)
-
COVID-19
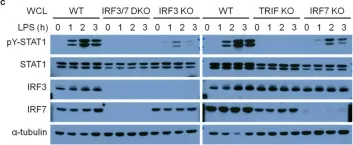
In Front Immunol on 7 May 2020 by Sin, W. X., Yeong, J. P., et al.
Fig.7.C

-
WB
-
Mus musculus (House mouse)
Collected and cropped from Front Immunol by CiteAb, provided under a CC-BY license
Image 1 of 2
In Sci Rep on 25 March 2019 by Mace, T. A., Ware, M. B., et al.
Fig.6.C

-
WB
-
Collected and cropped from Sci Rep by CiteAb, provided under a CC-BY license
Image 1 of 2