Ribosome quality control (RQC) resolves collided ribosomes, thus preventing their cytotoxic effects. The chemotherapeutic agent 5-Fluorouracil (5FU) is best known for its misincorporation into DNA and inhibition of thymidylate synthase. However, while a major determinant of 5FU's anticancer activity is its misincorporation into RNAs, the mechanisms by which cancer cells overcome the RNA-dependent 5FU toxicity remain ill-defined. Here, we report a role for RQC in mitigating the cytotoxic effects of 5FU. We show that 5FU treatment results in rapid induction of the mTOR signalling pathway, enhanced rate of mRNA translation initiation, and increased ribosome collisions. Consistently, a defective RQC exacerbates the 5FU-induced cell death, which is mitigated by blocking mTOR pathway or mRNA translation initiation. Furthermore, 5FU treatment enhances the expression of the key RQC factors ZNF598 and GIGYF2 via an mTOR-dependent post-translational mechanism. This adaptation likely mitigates the cytotoxic consequences of increased ribosome collisions upon 5FU treatment.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Product Citations: 44
In Nucleic Acids Research on 11 November 2024 by Chatterjee, S., Naeli, P., et al.
-
Biochemistry and Molecular biology
Repression of mRNA translation initiation by GIGYF1 via disrupting the eIF3-eIF4G1 interaction.
In Science Advances on 19 July 2024 by Choi, J. H., Luo, J., et al.
Viruses can selectively repress the translation of mRNAs involved in the antiviral response. RNA viruses exploit the Grb10-interacting GYF (glycine-tyrosine-phenylalanine) proteins 2 (GIGYF2) and eukaryotic translation initiation factor 4E (eIF4E) homologous protein 4EHP to selectively repress the translation of transcripts such as Ifnb1, which encodes the antiviral cytokine interferon-β (IFN-β). Herein, we reveal that GIGYF1, a paralog of GIGYF2, robustly represses cellular mRNA translation through a distinct 4EHP-independent mechanism. Upon recruitment to a target mRNA, GIGYF1 binds to subunits of eukaryotic translation initiation factor 3 (eIF3) at the eIF3-eIF4G1 interaction interface. This interaction disrupts the eIF3 binding to eIF4G1, resulting in transcript-specific translational repression. Depletion of GIGYF1 induces a robust immune response by derepressing IFN-β production. Our study highlights a unique mechanism of translational regulation by GIGYF1 that involves sequestering eIF3 and abrogating its binding to eIF4G1. This mechanism has profound implications for the host response to viral infections.
-
Biochemistry and Molecular biology
-
Genetics
Preprint on BioRxiv : the Preprint Server for Biology on 26 December 2023 by Chatterjee, S., Naeli, P., et al.
Translation of aberrant or damaged mRNAs results in ribosome stalling and collisions. The Ribosome Quality Control (RQC) mechanism detects collided ribosomes and removes aberrant mRNAs and nascent peptides, thus preventing their cytotoxic effects. Conversely, excessive or unresolved ribosome collisions can induce apoptosis. 5-Fluorouracil (5FU) forms the backbone of standard-of-care chemotherapeutic regimens for several types of cancer. Although best known for its incorporation into DNA and inhibition of thymidylate synthase, a major determinant of 5FU’s anticancer activity is its incorporation into RNAs. Nevertheless, the mechanism(s) underlying RNA-dependent 5FU cytotoxicity and the cellular response to its impact on RNA metabolism remain unclear. Here, we report a key role for RQC in mitigating the cytotoxic effects of 5FU-induced dysregulation of mRNA translation. We show that acute 5FU treatment results in the rapid induction of the mTOR signalling pathway, an enhanced rate of mRNA translation initiation, and increased ribosome collisions that trigger RQC. We also found that RQC deficiency, caused by the depletion of ZNF598, results in increased 5FU-induced cell death, a phenotype that is reversed by inhibition of mTOR or repression of mRNA translation initiation. Importantly, 5FU treatment enhances the expression of key RQC factors, including ZNF598 and GIGYF2, via an mTOR-dependent post-translational regulation mechanism. This acute adaptation likely mitigates the cytotoxic consequences of increased ribosome collisions upon 5FU treatment. Overall, our data indicate a heretofore unknown mTOR-dependent mechanism that augments the RQC process, mitigating the cytotoxicity of 5FU and undermining its anticancer efficacy.
Repression of mRNA translation initiation by GIGYF1 via blocking the eIF3-eIF4G1 interaction
Preprint on BioRxiv : the Preprint Server for Biology on 15 October 2023 by Choi, J., Luo, J., et al.
Summary Viruses commonly interfere with the function of the eukaryotic translation initiation factor 4G1 (eIF4G1), a pivotal factor in the recruitment of the eIF3 complex and ribosome to the mRNA. This results in the inhibition of general host protein synthesis and redirecting ribosomes toward viral mRNAs. Certain viruses also selectively repress the translation of mRNAs involved in the host antiviral response. GIGYF2 and its interacting cap-binding protein 4EHP enable the transcript-specific repression of mRNA translation mediated by microRNAs and RNA-binding proteins (RBPs). RNA viruses, such as SARS-CoV-2, exploit the GIGYF2/4EHP complex to selectively repress the translation of transcripts such as Ifnb1 mRNA, which encodes the antiviral cytokine Interferon β (IFN-β). Herein, we reveal that GIGYF1, a paralogue of GIGYF2, robustly represses cellular mRNA translation through a distinct mechanism independent of 4EHP. Upon recruitment to a target mRNA by RBPs, the C-terminal region of GIGYF1 binds to subunits of eIF3 at the interaction interface of eIF3-eIF4G1. This disrupts binding of eIF3 to eIF4G1, resulting in mRNA-specific translational repression. This mechanism exerts profound influences on the host cell’s response to viral infection. Depletion of GIGYF1 induces a robust immune response by derepressing Ifnb1 mRNA translation. Overall, our study highlights a unique mechanism of translational regulation by GIGYF1 that involves sequestering eIF3 and abrogating its binding to eIF4G1. This mechanism can be utilized by RBPs that interact with GIGYF1 to specifically repress the translation of their target mRNAs, significantly affecting critical biological processes, including host-pathogen interactions.
-
Biochemistry and Molecular biology
-
Genetics
N-myc-Mediated Translation Control Is a Therapeutic Vulnerability in Medulloblastoma.
In Cancer Research on 4 January 2023 by Kuzuoğlu-Öztürk, D., Aksoy, O., et al.
Deregulation of neuroblastoma-derived myc (N-myc) is a leading cause of malignant brain tumors in children. To target N-myc-driven medulloblastoma, most research has focused on identifying genomic alterations or on the analysis of the medulloblastoma transcriptome. Here, we have broadly characterized the translatome of medulloblastoma and shown that N-myc unexpectedly drives selective translation of transcripts that promote protein homeostasis. Cancer cells are constantly exposed to proteotoxic stress associated with alterations in protein production or folding. It remains poorly understood how cancers cope with proteotoxic stress to promote their growth. Here, our data revealed that N-myc regulates the expression of specific components (∼5%) of the protein folding machinery at the translational level through the major cap binding protein, eukaryotic initiation factor eIF4E. Reducing eIF4E levels in mouse models of medulloblastoma blocked tumorigenesis. Importantly, targeting Hsp70, a protein folding chaperone translationally regulated by N-myc, suppressed tumor growth in mouse and human medulloblastoma xenograft models. These findings reveal a previously hidden molecular program that promotes medulloblastoma formation and identify new therapies that may have impact in the clinic.
Translatome analysis in medulloblastoma shows that N-myc drives selective translation of transcripts that promote protein homeostasis and that represent new therapeutic vulnerabilities.
©2022 American Association for Cancer Research.
-
WB
-
Mus musculus (House mouse)
-
Biochemistry and Molecular biology
-
Cancer Research
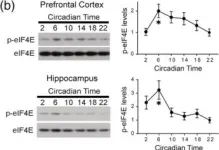
In Eur J Neurosci on 1 July 2022 by Liu, D., Li, J., et al.
Fig.2.B

-
WB
-
Collected and cropped from Eur J Neurosci by CiteAb, provided under a CC-BY license
Image 1 of 10
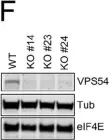
In Cancer Res Commun on 1 May 2022 by Marastoni, S., Madariaga, A., et al.
Fig.2.F

-
WB
-
Collected and cropped from Cancer Res Commun by CiteAb, provided under a CC-BY license
Image 1 of 10
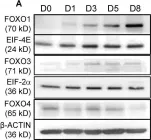
In Cells on 26 March 2022 by Zhong, Q., Liu, Y., et al.
Fig.1.A

-
WB
-
Collected and cropped from Cells by CiteAb, provided under a CC-BY license
Image 1 of 10
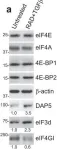
In Nat Commun on 30 November 2021 by Volta, V., Pérez-Baos, S., et al.
Fig.7.A

-
WB
-
Collected and cropped from Nat Commun by CiteAb, provided under a CC-BY license
Image 1 of 10
In Nat Commun on 30 November 2021 by Volta, V., Pérez-Baos, S., et al.
Fig.7.G

-
WB
-
Collected and cropped from Nat Commun by CiteAb, provided under a CC-BY license
Image 1 of 10
In Nat Commun on 30 November 2021 by Volta, V., Pérez-Baos, S., et al.
Fig.7.E

-
WB
-
Collected and cropped from Nat Commun by CiteAb, provided under a CC-BY license
Image 1 of 10
In Nat Commun on 30 November 2021 by Volta, V., Pérez-Baos, S., et al.
Fig.7.B

-
WB
-
Collected and cropped from Nat Commun by CiteAb, provided under a CC-BY license
Image 1 of 10
In Cells on 26 November 2021 by Fang, K., Liu, D., et al.
Fig.4.B

-
WB
-
Collected and cropped from Cells by CiteAb, provided under a CC-BY license
Image 1 of 10
In Sci Rep on 18 December 2015 by Smith, K. A., Zhou, B., et al.
Fig.2.A

-
WB
-
Collected and cropped from Sci Rep by CiteAb, provided under a CC-BY license
Image 1 of 10
In Elife on 20 May 2014 by Dixon, S. J., Patel, D. N., et al.
Fig.4.A

-
WB
-
Collected and cropped from Elife by CiteAb, provided under a CC-BY license
Image 1 of 10