The Covid-19 infection outbreak led to a global epidemic, and although several vaccines have been developed, the appearance of mutations has allowed the virus to evade the immune response. Added to this is the existing risk of the appearance of new emerging viruses. Therefore, it is necessary to explore novel antiviral therapies. Here, we investigate the potential in vitro of plant extracts to modulate cellular stress and inhibit murine hepatitis virus (MHV)-A59 replication. L929 cells were treated with P2Et (Caesalpinia spinosa) and Anamu SC (Petiveria alliacea) plant extracts during infection and virus production, ROS (reactive oxygen species), UPR (unfolded protein response), and autophagy were assessed. P2Et inhibited virus replication and attenuated both ROS production and UPR activation induced during infection. In contrast, the sustained presence of Anamu SC during viral adsorption and replication was required to inhibit viral infection, tending to induce pro-oxidant effects, and increasing UPR gene expression. Notably, the loss of the PERK protein resulted in a slight decrease in virus yield, suggesting a potential involvement of this UPR pathway during replication. Intriguingly, both extracts either maintained or increased the calreticulin surface exposure induced during infection. In conclusion, our findings highlight the development of antiviral natural plant extracts that differentially modulate cellular stress.
© 2023 The Authors.
Product Citations: 36
Plant extracts modulate cellular stress to inhibit replication of mouse Coronavirus MHV-A59.
In Heliyon on 15 January 2024 by Prieto, K., Arévalo, C., et al.
-
WB
-
Mus musculus (House mouse)
Murine leukemia virus infection of non-dividing dendritic cells is dependent on nucleoporins.
In PLoS Pathogens on 1 January 2024 by Salas-Briceno, K., Zhao, W., et al.
Retroviral reverse transcription starts within the capsid and uncoating and reverse transcription are mutually dependent. There is still debate regarding the timing and cellular location of HIV's uncoating and reverse transcription and whether it occurs solely in the cytoplasm, nucleus or both. HIV can infect non-dividing cells because there is active transport of the preintegration complex (PIC) across the nuclear membrane, but Murine Leukemia Virus (MLV) is thought to depend on cell division for replication and whether MLV uncoating and reverse transcription is solely cytoplasmic has not been studied. Here, we used NIH3T3 and primary mouse dendritic cells to determine where the different stages of reverse transcription occur and whether cell division is needed for nuclear entry. Our data strongly suggest that in both NIH3T3 cells and dendritic cells (DCs), the initial step of reverse transcription occurs in the cytoplasm. However, we detected MLV RNA/DNA hybrid intermediates in the nucleus of dividing NIH3T3 cells and non-dividing DCs, suggesting that reverse transcription can continue after nuclear entry. We also confirmed that the MLV PIC requires cell division to enter the nucleus of NIH3T3 cells. In contrast, we show that MLV can infect non-dividing primary DCs, although integration of MLV DNA in DCs still required the viral p12 protein. Knockdown of several nuclear pore proteins dramatically reduced the appearance of integrated MLV DNA in DCs but not NIH3T3 cells. Additionally, MLV capsid associated with the nuclear pore proteins NUP358 and NUP62 during infection. These findings suggest that simple retroviruses, like the complex retrovirus HIV, gain nuclear entry by traversing the nuclear pore complex in non-mitotic cells.
Copyright: © 2024 Salas-Briceno et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
-
Cancer Research
-
Immunology and Microbiology
In The Journal of Investigative Dermatology on 1 September 2023 by Eriksson, I., Vainikka, L., et al.
Lysosomes are central in cell homeostasis and participate in macromolecular degradation, plasma membrane repair, exosome release, cell adhesion/migration, and apoptosis. In cancer, alterations in lysosomal function and spatial distribution may facilitate disease progression. In this study, we show enhanced lysosomal activity in malignant melanoma cells compared with that in normal human melanocytes. Most lysosomes show perinuclear location in melanocytes, while they are more dispersed in melanoma, with retained proteolytic activity and low pH also in the peripheral population. Rab7a expression is lower in melanoma cells than in melanocytes, and by increasing Rab7a, lysosomes are relocated to the perinuclear region in melanoma. Exposure to the lysosome-destabilizing drug L-leucyl-L-leucine methyl ester causes higher damage in the perinuclear subset of lysosomes in melanomas, whereas differences in subpopulation susceptibility cannot be found in melanocytes. Interestingly, melanoma cells recruit the endosomal sorting complex required for transport-III core protein CHMP4B, involved in lysosomal membrane repair, rather than initiate lysophagy. However, when the perinuclear lysosomal position is promoted by Rab7a overexpression or kinesore treatment, lysophagy is increased. In addition, Rab7a overexpression is accompanied by reduced migration capacity. Taken together, the study emphasizes that alterations in lysosomal properties facilitate the malignant phenotype and declares the targeting of lysosomal function as a future therapeutic approach.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
-
Cancer Research
-
Cell Biology
Murine leukemia virus can infect non-dividing cells
Preprint on BioRxiv : the Preprint Server for Biology on 30 August 2023 by Salas-Briceno, K., Zhao, W., et al.
Retroviral reverse transcription starts within the capsid and uncoating and reverse transcription are mutually dependent. There is still debate regarding the timing and cellular location of HIV’s uncoating and reverse transcription and whether it occurs solely in the cytoplasm, nucleus or both. HIV can infect non-dividing cells because there is active transport of the preintegration complex (PIC) across the nuclear membrane, but Murine Leukemia Virus (MLV) is thought to depend on cell division for replication and whether MLV uncoating and reverse transcription is solely cytoplasmic has not been studied. Here, we used NIH3T3 and primary mouse dendritic cells to determine where the different stages of reverse transcription occur and whether cell division is needed for nuclear entry. Our data strongly suggest that in both NIH3T3 cells and dendritic cells (DCs), the initial step of reverse transcription occurs in the cytoplasm. However, we detected MLV RNA/DNA hybrid intermediates in the nucleus of dividing NIH3T3 cells and non-dividing DCs, suggesting that reverse transcription can continue after nuclear entry. We also found that the MLV PIC requires cell division to enter the nucleus of NIH3T3 cells. In contrast, we show that MLV can infect non-dividing primary DCs, although integration of MLV DNA in DCs still required the viral p12 protein. Knockdown of several nuclear pore proteins dramatically reduced the appearance of integrated MLV DNA in DCs but not NIH3T3 cells. Additionally, MLV capsid associates with the nuclear pore proteins NUP358 and NUP62 during infection. These findings suggest that simple retroviruses, like HIV, gain nuclear entry by traversing the nuclear pore complex in non-mitotic cells. Author Summary It is widely believed that gammaretroviruses like MLV require cell division to achieve nuclear entry and complete their replication. We show here that while this is true for rapidly dividing tissue culture cells, in quiescent cells like dendritic cells, the natural targets of MLV infection, the virus establishes infection without cell division. These studies show that the requirements for retrovirus infection depend on the cell type.
-
Cancer Research
-
Immunology and Microbiology
Functional glycoproteomics by integrated network assembly and partitioning
Preprint on BioRxiv : the Preprint Server for Biology on 14 June 2023 by Griffin, M. E., Thompson, J. W., et al.
SUMMARY The post-translational modification (PTM) of proteins by O-linked β- N -acetyl-D-glucosamine (O-GlcNAcylation) is widespread across the proteome during the lifespan of all multicellular organisms. However, nearly all functional studies have focused on individual protein modifications, overlooking the multitude of simultaneous O-GlcNAcylation events that work together to coordinate cellular activities. Here, we describe N etworking of I nteractors and S ubstrat E s (NISE), a novel, systems-level approach to rapidly and comprehensively monitor O-GlcNAcylation across the proteome. Our method integrates affinity purification-mass spectrometry (AP-MS) and site-specific chemoproteomic technologies with network generation and unsupervised partitioning to connect potential upstream regulators with downstream targets of O-GlcNAcylation. The resulting network provides a data-rich framework that reveals both conserved activities of O-GlcNAcylation such as epigenetic regulation as well as tissue-specific functions like synaptic morphology. Beyond O-GlcNAc, this holistic and unbiased systems-level approach provides a broadly applicable framework to study PTMs and discover their diverse roles in specific cell types and biological states.
-
Homo sapiens (Human)
In Cell Death Dis on 13 August 2019 by Turkieh, A., Porouchani, S., et al.
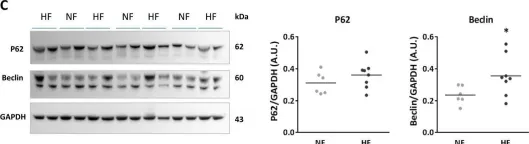
Fig.6.C

-
WB
-
Collected and cropped from Cell Death Dis by CiteAb, provided under a CC-BY license
Image 1 of 2
In Front Mol Neurosci on 20 October 2018 by Cuvelier, E., Méquinion, M., et al.
Fig.6.A

-
WB
-
Collected and cropped from Front Mol Neurosci by CiteAb, provided under a CC-BY license
Image 1 of 2