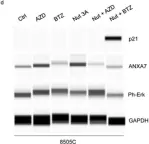
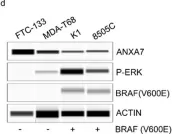
Thyroid cancer is the most common endocrine malignancy in the United States, with an overall favorable prognosis. However, some patients experience poor outcomes due to the development of resistance to conventional therapies. Genetic alterations, including mutations in BRAF, Met, and p53, play critical roles in thyroid cancer progression, with the BRAF V600E mutation detected in over 60% of cases. This study investigates the tumor-suppressive role of Annexin A7 (ANXA7) in thyroid cancer, focusing on its potential impact on tumor behavior and therapeutic response. Our analysis, which included RNA sequencing and protein profiling, revealed reduced ANXA7 expression in thyroid cancer cells, particularly in those harboring the BRAF V600E mutation. Upon treatment with inhibitors targeting BRAF and MEK, ANXA7 expression increased, leading to reduced phosphorylation of ERK and activation of apoptotic pathways. Additionally, we identified the cyclin-dependent kinase inhibitor p21 as a key player in modulating resistance to BRAF inhibitors. Combination therapies aimed at concurrently increasing p21 and ANXA7 levels resulted in a marked enhancement of apoptosis. These findings suggest a previously uncharacterized regulatory network involving the ANXA7/p21/BRAF/MAPK/p53 axis, which may contribute to drug resistance in thyroid cancer. This study provides new insights into overcoming resistance to BRAF and MAPK inhibitors, with implications for treating thyroid cancer and potentially other BRAF-mutant tumors. Future efforts will focus on high-throughput screening approaches to explore ANXA7-targeted therapeutic strategies for thyroid cancer.
Product Citations: 6
In International Journal of Molecular Sciences on 9 December 2024 by Bera, A., Radhakrishnan, S., et al.
-
SW - Size
-
Homo sapiens (Human)
-
Cancer Research
-
Endocrinology and Physiology
In Biochemical Pharmacology on 1 August 2019 by Aareskjold, E., Grindheim, A. K., et al.
Besides altering its own expression during cell transformation, Annexin A2 is upregulated during the progression of many cancer types and also plays key roles during viral infection and multiplication. Consequently, there has been great interest in Annexin A2 as a potential drug target. The successful design of efficient in vivo delivery systems constitutes an obstacle in full exploitation of antisense and RNA-cleaving technologies for the knock-down of specific targets. Efficiency is dependent on the method of delivery and accessibility of the target. Here, hairpin ribozymes and an antisense RNA against rat annexin A2 mRNA were tested for their efficiencies in a T7-driven coupled transcription/translation system. The most efficient ribozyme and antisense RNA were subsequently inserted into a retroviral vector under the control of a tRNA promoter, in a cassette inserted between retroviral Long Terminal Repeats for stable insertion into host DNA. The Phoenix package system based on defective retroviruses was used for virus-mediated gene transfer into PC12 cells. Cells infected with the ribozyme-containing particles died shortly after infection. However, the same ribozyme showed a very high catalytic effect in vitro in cell lysates, explained by its loose hinge helix 2 region. This principle can be transferred to other ribozymes, such as those designed to cleave the guide RNA in the CRISPR/Cas9 technology, as well as to target specific viral RNAs. Interestingly, efficient down-regulation of the expression of Annexin A2 by the antisense RNA resulted in up-regulation of Annexin A7 as a compensatory effect after several cell passages. Indeed, compensatory effects have previously been observed during gene knock-out, but not during knock-down of protein expression. This highlights the problems in interpreting the phenotypic effects of knocking down the expression of a protein. In addition, these data are highly relevant when considering the effects of the CRISPR/Cas9 approach.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
-
Biochemistry and Molecular biology
-
Genetics
In PLoS ONE on 16 October 2018 by Leighton, X., Bera, A., et al.
Annexin A7 (ANXA7) is a member of the multifunctional calcium or phospholipid-binding annexin gene family. While low levels of ANXA7 are associated with aggressive types of cancer, the clinical impact of ANXA7 in prostate cancer remains unclear. Tissue microarrays (TMA) have revealed several new molecular markers in human tumors. Herein, we have identified the prognostic impact of ANXA7 in a prostate cancer using a tissue microarray containing 637 different specimens.
The patients were diagnosed with prostate cancer and long-term follow-up information on progression (median 5.3 years), tumor-specific and overall survival data (median 5.9 years) were available. Expression of Ki67, Bcl-2, p53, CD-10 (neutral endopeptidase), syndecan-1 (CD-138) and ANXA7 were analyzed by immunohistochemistry.
A bimodal distribution of ANXA7 was observed. Tumors expressing either high or no ANXA7 were found to be associated with poor prognosis. However, ANXA7 at an optimal level, in between high and no ANXA7 expression, had a better prognosis. This correlated with low Ki67, Bcl-2, p53 and high syndecan-1 which are known predictors of early recurrence. At Gleason grade 3, ANXA7 is an independent predictor of poor overall survival with a p-value of 0.003. Neoadjuvant hormonal therapy, which is known to be associated with overexpression of Bcl-2 and inhibition of Ki67 LI and CD-10, was found to be associated with under-expression of ANXA7.
The results of this TMA study identified ANXA7 as a new prognostic factor and indicates a bimodal correlation to tumor progression.
-
IHC
-
Homo sapiens (Human)
-
Cancer Research
Systematic Functional Annotation of Somatic Mutations in Cancer.
In Cancer Cell on 12 March 2018 by Ng, P. K., Li, J., et al.
The functional impact of the vast majority of cancer somatic mutations remains unknown, representing a critical knowledge gap for implementing precision oncology. Here, we report the development of a moderate-throughput functional genomic platform consisting of efficient mutant generation, sensitive viability assays using two growth factor-dependent cell models, and functional proteomic profiling of signaling effects for select aberrations. We apply the platform to annotate >1,000 genomic aberrations, including gene amplifications, point mutations, indels, and gene fusions, potentially doubling the number of driver mutations characterized in clinically actionable genes. Further, the platform is sufficiently sensitive to identify weak drivers. Our data are accessible through a user-friendly, public data portal. Our study will facilitate biomarker discovery, prediction algorithm improvement, and drug development.
Copyright © 2018 Elsevier Inc. All rights reserved.
-
ICC
-
Homo sapiens (Human)
-
Cancer Research
Context Specificity in Causal Signaling Networks Revealed by Phosphoprotein Profiling.
In Cell Systems on 25 January 2017 by Hill, S. M., Nesser, N. K., et al.
Signaling networks downstream of receptor tyrosine kinases are among the most extensively studied biological networks, but new approaches are needed to elucidate causal relationships between network components and understand how such relationships are influenced by biological context and disease. Here, we investigate the context specificity of signaling networks within a causal conceptual framework using reverse-phase protein array time-course assays and network analysis approaches. We focus on a well-defined set of signaling proteins profiled under inhibition with five kinase inhibitors in 32 contexts: four breast cancer cell lines (MCF7, UACC812, BT20, and BT549) under eight stimulus conditions. The data, spanning multiple pathways and comprising ∼70,000 phosphoprotein and ∼260,000 protein measurements, provide a wealth of testable, context-specific hypotheses, several of which we experimentally validate. Furthermore, the data provide a unique resource for computational methods development, permitting empirical assessment of causal network learning in a complex, mammalian setting.Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
-
Ab Array
-
Homo sapiens (Human)
In Int J Mol Sci on 9 December 2024 by Bera, A., Radhakrishnan, S., et al.
Fig.7.D

-
SW - Size
-
Homo sapiens (Human)
Collected and cropped from Int J Mol Sci by CiteAb, provided under a CC-BY license
Image 1 of 2
In Int J Mol Sci on 9 December 2024 by Bera, A., Radhakrishnan, S., et al.
Fig.2.D

-
SW - Size
-
Homo sapiens (Human)
Collected and cropped from Int J Mol Sci by CiteAb, provided under a CC-BY license
Image 1 of 2