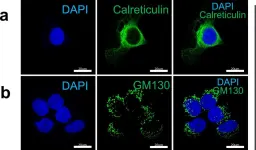
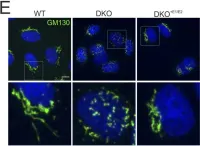
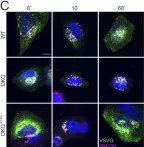
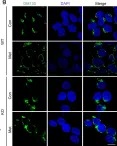
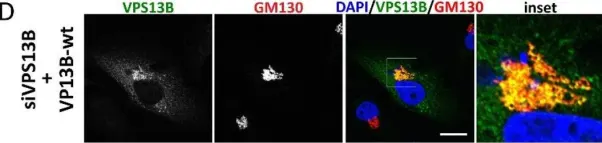
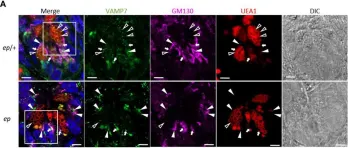
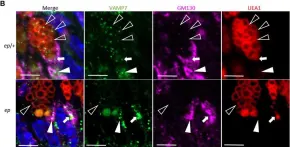
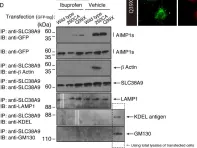
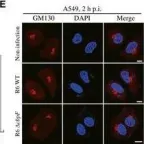
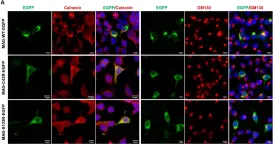
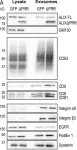
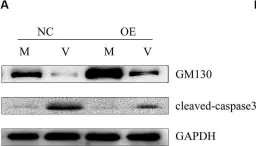
κB-Ras/RalGAP complexes limit the activity of Ral GTPases, which function in EGFR/Ras signaling. RalGAP expression is down-regulated in pancreatic cancer; however, the role of RalGAP and Ral GTPases in tumor development in vivo remained unclear. Here, we show that pancreatic RalGAPβ deficiency alone is sufficient to induce inflammation and neoplasia in vivo. We identify that this phenotype is triggered by disturbance of the secretory pathway and polarized exocytosis in acinar cells, demonstrating that RalGAP complexes uphold spatial control of Ral activity. We furthermore show that RALGAPβ deficiency results in defective primary cilium assembly, a process required for efficient acinar regeneration upon inflammation. Only primary cilium formation depends on κB-Ras proteins, suggesting that κB-Ras proteins are not essential for all RalGAP complex-controlled processes. In combination with an oncogenic KRAS G12D mutation, RalGAPβ deficiency leads to a dramatic shortening of tumor latency and median survival. Our results highlight an important role of RalGAP/Ral signaling in upholding acinar cell identity and preventing pancreatic cancer development.
© 2025 Apken et al.
Product Citations: 1,024
RalGAP complexes control secretion and primary cilia in pancreatic disease.
In Life Science Alliance on 1 August 2025 by Apken, L. H., Barz, H., et al.
In Scientific Reports on 3 June 2025 by Lei, R., da Silva, T. B., et al.
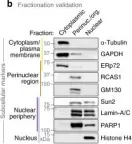
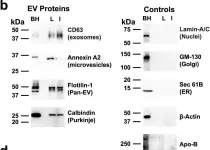
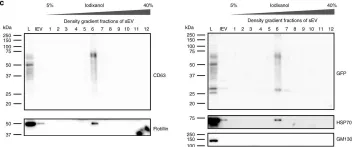
Studies have shown that mesenchymal stromal cells (MSCs) could secrete a variety of bioactive particles, including small extracellular vesicles (sEVs), that might be a key intermediate to the beneficial paracrine effects of MSC therapy. In this study, we harvested the conditioned medium (CM) from human bone marrow mesenchymal stromal cells (hBM-MSCs) and normal human dermal fibroblasts (NHDFs), fractionated into the sEV fraction and non-small extracellular vesicle (NsEV) fraction, and compared their functions on NHDF migration and proliferation-key processes in wound healing, coupled with transcriptomic analysis through mRNA sequencing to assess gene expression changes in the recipient NHDFs. Our findings show that sEV, NsEV and CM from hBM-MSCs had an overall promotive effect on migration behaviour of NHDFs, but MSC-sEV surpassed the effects of other MSC secretome fractions and their NHDF counterparts (HDF-sEV). Gene ontology analysis revealed enrichment of pathways related to migration, and significant changes in genes within the regulation of proliferation pathway. Our study provides referential significance for the choice of cellular secretome fractions to be used in wound healing studies and insights into the effects of MSC secretome in the migration and proliferation of healthy fibroblasts, besides their effects on gene expression changes.
© 2025. The Author(s).
In Bio-protocol on 20 May 2025 by Ishida, K. & Morita, E.
Orthoflavivirus is an enveloped, positive-stranded RNA virus that buds into the endoplasmic reticulum (ER) lumen. The budded virus particles are subsequently transported to the Golgi apparatus and secreted into the extracellular environment via the conventional secretion pathway. In this protocol, we describe a method for monitoring the secretion of Orthoflavivirus particles from the ER. To visualize intracellular membrane trafficking, we combine two distinct imaging techniques: the retention using selective hooks (RUSH) system and the split green fluorescent protein (GFP) system. In this approach, GFP11, a peptide tag fused to prME, the outer coat structural protein of Japanese encephalitis virus particles, was co-expressed in HeLa cells along with two additional components: GFP1-10 fused to a streptavidin-binding peptide and a hook construct consisting of streptavidin fused to the ER retention sequence KDEL. Time-lapse imaging was performed after the addition of biotin, which releases the captured GFP-labeled subviral particles from the ER. This method enables synchronized visualization of intracellular subviral particle trafficking and serves as a valuable tool for analyzing the maturation process of Orthoflavivirus particles within cells. Key features • Synchronized intracellular movement of Orthoflavivirus particles is visualized by the retention using selective hooks (RUSH) system. • Split GFP system is used to label viral particles. • This protocol has broader applications in investigating the transport of secretory proteins, especially those that are challenging to tag with full-length fluorescent proteins.
©Copyright : © 2025 The Authors; This is an open access article under the CC BY-NC license.
SARS-CoV-2 ORF3a drives dynamic dense body formation for optimal viral infectivity.
In Nature Communications on 12 May 2025 by Hartmann, S., Radochonski, L., et al.
SARS-CoV-2 hijacks multiple organelles for virion assembly, of which the mechanisms have not been fully understood. Here, we identified a SARS-CoV-2-driven membrane structure named the 3a dense body (3DB). 3DBs are unusual electron-dense and dynamic structures driven by the accessory protein ORF3a via remodeling a specific subset of the trans-Golgi network (TGN) and early endosomal membrane. 3DB formation is conserved in related bat and pangolin coronaviruses but was lost during the evolution to SARS-CoV. During SARS-CoV-2 infection, 3DB recruits the viral structural proteins spike (S) and membrane (M) and undergoes dynamic fusion/fission to maintain the optimal unprocessed-to-processed ratio of S on assembled virions. Disruption of 3DB formation resulted in virions assembled with an abnormal S processing rate, leading to a dramatic reduction in viral entry efficiency. Our study uncovers the crucial role of 3DB in maintaining maximal SARS-CoV-2 infectivity and highlights its potential as a target for COVID-19 prophylactics and therapeutics.
© 2025. The Author(s).
-
COVID-19
-
Immunology and Microbiology
STING agonist-based ER-targeting molecules boost antigen cross-presentation.
In Nature on 1 May 2025 by Wang, X., Huang, Z., et al.
CD8+ T cell immune responses are critical for combating infectious diseases and tumours1-3. Antigen cross-presentation, primarily occurring at the endoplasmic reticulum (ER) of dendritic cells, is essential for protein-based vaccines to induce CD8+ T cell responses4. Current efforts have focused on antigen delivery at the tissue and cellular levels, whereas subcellular delivery has been limited to facilitating antigen escape from lysosomes into the cytosol. In the absence of a small-sized high-affinity ER-targeting molecule, the importance of the 'last mile' from the cytosol to the ER remains elusive. Here we developed stimulator of interferon genes (STING) agonist-based ER-targeting molecules (SABER), which effectively deliver antigens to the ER and cluster key machinery in cross-presentation to form microreactors by folding the ER membrane. Conjugation of SABER to various antigens substantially enhances the induction of CD8+ T cell immune responses to tumour neoantigens and conserved viral epitopes, far exceeding that achieved by mixtures of antigens with STING agonists or conventional adjuvants. SABER also retains a potent adjuvant effect, effectively enhancing the ability of a SARS-CoV-2 subunit vaccine to induce broadly neutralizing antibodies. This study provides a high-affinity ER-targeting delivery system and vaccine adjuvant, demonstrating that precise subcellular delivery targeting the last mile of cross-presentation can lead to a qualitative leap.
© 2025. The Author(s).
-
Immunology and Microbiology
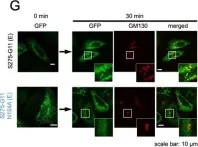
In Elife on 16 August 2024 by He, Q., Zhao, M. M., et al.
Fig.6.A

-
ICC-IF
-
Collected and cropped from Elife by CiteAb, provided under a CC-BY license
Image 1 of 100
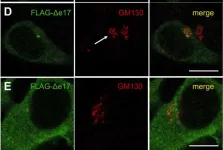
In Life Sci Alliance on 1 August 2024 by Veronese, M., Kallabis, S., et al.
Fig.6.E

-
ICC-IF
-
Homo sapiens (Human)
Collected and cropped from Life Sci Alliance by CiteAb, provided under a CC-BY license
Image 1 of 100
In Life Sci Alliance on 1 August 2024 by Veronese, M., Kallabis, S., et al.
Fig.6.C

-
ICC-IF
-
Homo sapiens (Human)
Collected and cropped from Life Sci Alliance by CiteAb, provided under a CC-BY license
Image 1 of 100
In Nat Commun on 27 February 2024 by Tu, Y., Yang, Q., et al.
Fig.3.G

-
ICC-IF
-
Collected and cropped from Nat Commun by CiteAb, provided under a CC-BY license
Image 1 of 100
In PLoS Pathog on 1 October 2023 by Ishida, K., Yagi, H., et al.
Fig.4.G

-
ICC-IF
-
Collected and cropped from PLoS Pathog by CiteAb, provided under a CC-BY license
Image 1 of 100
In Int J Biol Sci on 16 June 2023 by Ock, J., Wu, J., et al.
Fig.4.B

-
WB
-
Collected and cropped from Int J Biol Sci by CiteAb, provided under a CC-BY license
Image 1 of 100
In Sci Rep on 4 April 2023 by Kim, J. A., Park, C., et al.
Fig.1.C

-
WB
-
Homo sapiens (Human)
Collected and cropped from Sci Rep by CiteAb, provided under a CC-BY license
Image 1 of 100
In IBRO Neurosci Rep on 1 December 2022 by Nakamura, A., Ikeda, M., et al.
Fig.2.D

-
ICC-IF
-
Collected and cropped from IBRO Neurosci Rep by CiteAb, provided under a CC-BY license
Image 1 of 100
In IBRO Neurosci Rep on 1 December 2022 by Nakamura, A., Ikeda, M., et al.
Fig.3.C

-
ICC-IF
-
Collected and cropped from IBRO Neurosci Rep by CiteAb, provided under a CC-BY license
Image 1 of 100
In J Biol Chem on 1 August 2022 by Januário, Y. C., Eden, J., et al.
Fig.3.G

-
ICC-IF
-
Collected and cropped from J Biol Chem by CiteAb, provided under a CC-BY license
Image 1 of 100
In J Biol Chem on 1 August 2022 by Januário, Y. C., Eden, J., et al.
Fig.6.D

-
ICC-IF
-
Collected and cropped from J Biol Chem by CiteAb, provided under a CC-BY license
Image 1 of 100
In Sci Rep on 11 June 2022 by Zorn, M., Kühnisch, J., et al.
Fig.2.A

-
ICC-IF
-
Homo sapiens (Human)
Collected and cropped from Sci Rep by CiteAb, provided under a CC-BY license
Image 1 of 100
In Sci Rep on 11 June 2022 by Zorn, M., Kühnisch, J., et al.
Fig.2.D

-
ICC-IF
-
Homo sapiens (Human)
Collected and cropped from Sci Rep by CiteAb, provided under a CC-BY license
Image 1 of 100
In Nat Commun on 1 June 2022 by Byron, A., Griffith, B. G. C., et al.
Fig.2.B

-
WB
-
Mus musculus (House mouse)
Collected and cropped from Nat Commun by CiteAb, provided under a CC-BY license
Image 1 of 100
In Cell Death Dis on 20 December 2021 by Khan, S., Sbeity, M., et al.
Fig.1.C

-
ICC-IF
-
Collected and cropped from Cell Death Dis by CiteAb, provided under a CC-BY license
Image 1 of 100
In Nat Commun on 19 March 2021 by Zhang, Y., Varela, L., et al.
Fig.9.B

-
WB
-
Collected and cropped from Nat Commun by CiteAb, provided under a CC-BY license
Image 1 of 100
In Front Immunol on 24 November 2020 by Yu, J., He, X., et al.
Fig.6.A

-
WB
-
Collected and cropped from Front Immunol by CiteAb, provided under a CC-BY license
Image 1 of 100
In Front Immunol on 24 November 2020 by Yu, J., He, X., et al.
Fig.7.A

-
IHC-IF
-
Collected and cropped from Front Immunol by CiteAb, provided under a CC-BY license
Image 1 of 100
In Front Immunol on 24 November 2020 by Yu, J., He, X., et al.
Fig.7.B

-
IHC-IF
-
Collected and cropped from Front Immunol by CiteAb, provided under a CC-BY license
Image 1 of 100
In Medicines (Basel) on 6 May 2020 by Takeuchi, Y., Tanaka, M., et al.
Fig.9.D

-
ICC-IF
-
Collected and cropped from Medicines (Basel) by CiteAb, provided under a CC-BY license
Image 1 of 100
In EMBO Rep on 6 May 2020 by Shizukuishi, S., Ogawa, M., et al.
Fig.3.E

-
ICC-IF
-
Collected and cropped from EMBO Rep by CiteAb, provided under a CC-BY license
Image 1 of 100
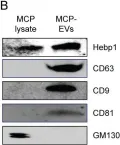
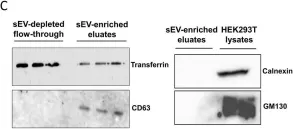
In Nat Commun on 29 April 2020 by Sung, B. H., von Lersner, A., et al.
Fig.2.C

-
WB
-
Collected and cropped from Nat Commun by CiteAb, provided under a CC-BY license
Image 1 of 100
In J Clin Med on 23 April 2020 by Santos, M., Damásio, J., et al.
Fig.3.A

-
ICC-IF
-
Homo sapiens (Human)
Collected and cropped from J Clin Med by CiteAb, provided under a CC-BY license
Image 1 of 100
In J Cell Biol on 2 March 2020 by Larios, J., Mercier, V., et al.
Fig.8.A

-
WB
-
Collected and cropped from J Cell Biol by CiteAb, provided under a CC-BY license
Image 1 of 100
In Front Mol Neurosci on 11 February 2020 by He, Q., Liu, H., et al.
Fig.6.A

-
WB
-
Collected and cropped from Front Mol Neurosci by CiteAb, provided under a CC-BY license
Image 1 of 100