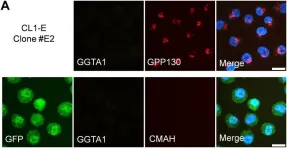
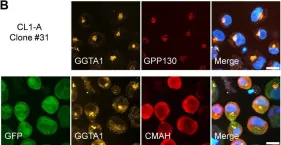
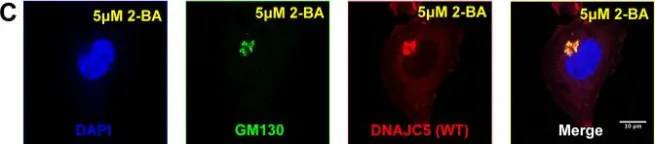
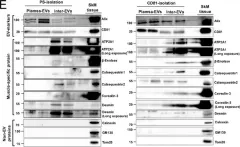
Stimulator of interferon genes (STING) triggers the type I interferon and inflammatory responses against a variety of DNA pathogens, which is essential to limiting viral infection and replication. STING activates the downstream kinase TBK1 at the trans-Golgi network (TGN) and is degraded at lysosomes through a process called lysosomal microautophagy. Impaired STING targeting to lysosomes results in the prolonged inflammatory signal, which may be associated with a variety of neurodegenerative and autoinflammatory diseases. Thus, development of methods to quantify STING degradation helps understand the mechanism of lysosomal microautophagy and its related diseases. Here we report a quantitative method to monitor STING degradation with two luciferases, firefly luciferase (FLuc) and Nanoluciferase (NLuc). The expression plasmid is composed of FLuc, a P2A self-cleavage site, and NLuc-tagged STING. FLuc intensity reflects the total amount of translated protein, serving as an internal control, while NLuc intensity corresponds to the amount of STING. Comparison of the NLuc/FLuc ratios at different time points after STING stimulation revealed the kinetics of decay of STING levels in live cells. This method should provide a useful complement to western blotting and fluorescence-activated cell sorter (FACS) analysis presently used to monitor STING degradation.Key words: innate immunity, STING, membrane traffic, lysosomal degradation, luciferase.
Product Citations: 480
A quantitative method to monitor STING degradation with dual-luciferase reporters.
In Cell Structure and Function on 20 May 2025 by Shoji, T., Sato, K., et al.
The proteasome maturation factor POMP moonlights as a stress-induced transcriptional regulator
Preprint on BioRxiv : the Preprint Server for Biology on 27 April 2025 by Giandomenico, S. L., Mueller, M., et al.
Summary Proteostasis - the maintenance of proteins at proper concentrations, conformations, and subcellular locations - is essential for cellular function and is governed by tightly regulated protein synthesis and degradation pathways. The Proteasome Maturation Protein (POMP) is a key chaperone involved in assembling the proteasome, the primary complex responsible for protein degradation. Despite the conserved role of POMP, its loss produces contrasting proteostatic effects in yeast and mammalian cells, pointing to additional, unexplored functions. In this study, we investigated the possibility that POMP plays a moonlighting role in proteostasis regulation. We discovered that upon proteasome disruption, POMP rapidly accumulates in the nucleolus in a manner dependent on HSF1 and reactive oxygen species (ROS). Proteomic analysis of POMP interactors revealed RNA processing factors and transcriptomic profiling showed that nucleolar POMP orchestrates a protective transcriptional program. Our findings indicate POMP is a built-in sensor and effector within the proteasome assembly pathway, capable of buffering disturbances in proteasome function through a novel, non-canonical role. Notably, this mechanism is developmentally controlled and active in neurodegenerative disease contexts. Abstract Figure
-
WB
-
Biochemistry and Molecular biology
In Frontiers in Molecular Biosciences on 15 April 2025 by Wiertelak, W., Pavlovskyi, A., et al.
Glycosylation is a vital post-translational modification involving the addition of sugars to proteins and lipids, facilitated by glycosyltransferases and dependent on nucleotide sugar donors like UDP-galactose (UDP-Gal). This study examines how disruptions in UDP-Gal synthesis affect protein-protein interactions critical for glycosylation. Using CRISPR/Cas9, we generated HEK293T cell lines lacking key enzymes of the Leloir pathway: UDP-galactose 4'-epimerase (GALE), galactose-1-phosphate uridylyltransferase (GALT), or both. The knockout of GALE led to a significant reduction in intracellular UDP-Gal levels and altered N-glycan profiles, indicating impaired galactosylation. Through the NanoBiT assay, we observed that knocking out GALE alone or both GALE and GALT diminished the ability of the UDP-Gal transporter SLC35A2 to form homomers and to interact with the beta-1,4-galactosyltransferase 1 (B4GALT1). These findings suggest that the nucleotide sugar availability and/or the presence of the corresponding enzymes in the cytoplasm influences the formation of protein complexes involved in glycosylation in the Golgi apparatus, potentially affecting the glycosylation process itself. Our study highlights the dynamic nature of the glycosylation machinery and suggests that the interactions between glycosylation proteins are responsive to changes in nucleotide sugar levels. This opens new avenues for understanding the mechanisms underlying glycosylation and for investigating congenital glycosylation disorders.
Copyright © 2025 Wiertelak, Pavlovskyi, Olczak and Maszczak-Seneczko.
-
Cell Biology
Preprint on BioRxiv : the Preprint Server for Biology on 14 April 2025 by Gao, J., Cong, C., et al.
ABSTRACT Many transmembrane (TM) receptors undergo essential post-translational modification in the Golgi prior to their delivery to the plasma membrane. Whether and how the passage and accompanied modification of these receptors across the Golgi is controlled remains unclear. Here, we show that leptin receptor overlapping transcript (LEPROT) and LEPROT-like 1 (LEPROTL1) regulate receptor activation by securing their sufficient Golgi retention. LEPROTs localize to cis and medial Golgi in a COPI-dependent manner. Deletion of LEPROTs in cells causes expedited release of receptors transiting through the Golgi, and leakage of some Golgi-enriched proteins into the endomembrane compartments. LEPROTs interact directly with COPI coats and simultaneously engage a variety of integral membrane proteins with relatively long TM domains at acidic pH. Loss of LEPROTs dysregulates receptor activity, including that of EGFR and TFRC, due to defective modification. Collectively, LEPROTs serve as a class of COPI adaptors for TM receptors, ensuring adequate preparation, which is vital for subsequent action on the plasma membrane.
-
Cell Biology
In Molecular Neurobiology on 10 April 2025 by Kadam, V., Wacker, M., et al.
Transmembrane L1 cell adhesion molecule (L1CAM) is widely used as a marker to enrich for neuron-derived extracellular vesicles (EVs), especially in plasma. However, this approach lacks sufficient robust validation. This study aimed to assess whether human biofluids are indeed enriched for EVs, particularly neuron-derived EVs, by L1CAM immunoaffinity, utilizing multiple sources (plasma, CSF, conditioned media from iPSC-derived neurons [iNCM]) and different methods (mass spectrometry [MS], nanoparticle tracking analysis [NTA]). Following a systematic multi-step validation approach, we confirmed isolation of generic EV populations using size-exclusion chromatography (SEC) and polymer-aided precipitation (PPT)-two most commonly applied EV isolation methods-from all sources. Neurofilament light (NfL) was detected in both CSF and blood-derived EVs, indicating their neuronal origin. However, L1CAM immunoprecipitation did not yield enrichment of L1CAM in EV fractions. Instead, it was predominantly found in its free-floating form. Additionally, MS-based proteomic analysis of CSF-derived EVs also did not show L1CAM enrichment. Our study validates EV isolation from diverse biofluid sources by several isolation approaches and confirms that some EV subpopulations in human biofluids are of neuronal origin. Thorough testing across multiple sources by different orthogonal methods, however, does not support L1CAM as a marker to reliably enrich for a specific subpopulation of EVs, particularly of neuronal origin.
© 2025. The Author(s).
-
WB
-
Neuroscience
-
Stem Cells and Developmental Biology
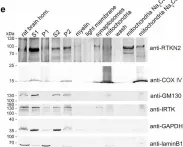
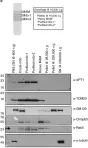
In Nat Commun on 10 March 2025 by Tröger, J., Dahlhaus, R., et al.
Fig.2.E

-
WB
-
Collected and cropped from Nat Commun by CiteAb, provided under a CC-BY license
Image 1 of 41
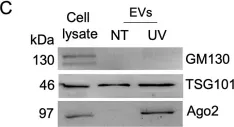
In Cell Commun Signal on 8 November 2024 by Salvi, V., Gaudenzi, C., et al.
Fig.2.C

-
WB
-
Collected and cropped from Cell Commun Signal by CiteAb, provided under a CC-BY license
Image 1 of 41
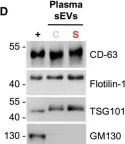
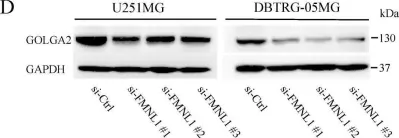
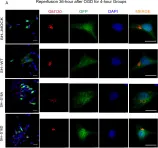
In Biol Res on 28 September 2024 by Sánchez-Rubio, M., Abarzua-Catalan, L., et al.
Fig.3.D

-
WB
-
Rattus norvegicus (Rat)
Collected and cropped from Biol Res by CiteAb, provided under a CC-BY license
Image 1 of 41
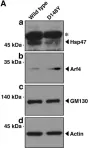
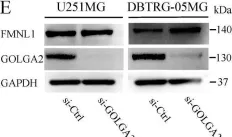
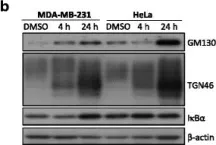
In Curr Issues Mol Biol on 5 February 2024 by Fukatsu, S., Okawa, M., et al.
Fig.2.A

-
WB
-
Collected and cropped from Curr Issues Mol Biol by CiteAb, provided under a CC-BY license
Image 1 of 41
In Front Bioeng Biotechnol on 7 March 2023 by Gupta, S., Shah, B., et al.
Fig.6.A

-
ICC-IF
-
Collected and cropped from Front Bioeng Biotechnol by CiteAb, provided under a CC-BY license
Image 1 of 41
In Front Bioeng Biotechnol on 7 March 2023 by Gupta, S., Shah, B., et al.
Fig.6.B

-
ICC-IF
-
Collected and cropped from Front Bioeng Biotechnol by CiteAb, provided under a CC-BY license
Image 1 of 41
In Elife on 10 January 2023 by Wu, S., Hernandez Villegas, N. C., et al.
Fig.3.C

-
IHC-IF
-
Collected and cropped from Elife by CiteAb, provided under a CC-BY license
Image 1 of 41
In PNAS Nexus on 1 September 2022 by Watanabe, S., Sudo, Y., et al.
Fig.4.E

-
WB
-
Collected and cropped from PNAS Nexus by CiteAb, provided under a CC-BY license
Image 1 of 41
In Int J Mol Sci on 2 August 2022 by Goto, A., Egawa, D., et al.
Fig.3.A

-
ICC-IF
-
Collected and cropped from Int J Mol Sci by CiteAb, provided under a CC-BY license
Image 1 of 41
In Sci Rep on 17 March 2022 by Husein, D., Alamoudi, A., et al.
Fig.6.B

-
WB
-
Collected and cropped from Sci Rep by CiteAb, provided under a CC-BY license
Image 1 of 41
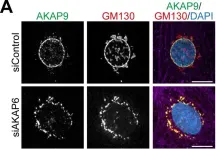
In Elife on 12 November 2021 by Liu, X. M., Ma, L., et al.
Fig.4.D

-
WB
-
Collected and cropped from Elife by CiteAb, provided under a CC-BY license
Image 1 of 41
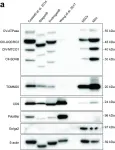
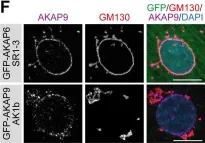
In Front Mol Neurosci on 24 August 2021 by Sampieri, L., Funes Chabán, M., et al.
Fig.1.F

-
WB
-
Collected and cropped from Front Mol Neurosci by CiteAb, provided under a CC-BY license
Image 1 of 41
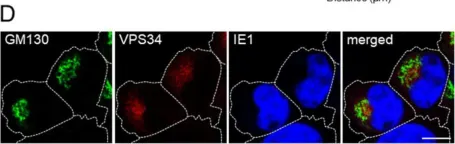
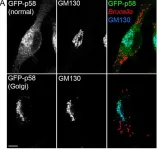
In Life (Basel) on 22 August 2021 by Marcelić, M., Mahmutefendić Lučin, H., et al.
Fig.1.D

-
ICC-IF
-
Mus musculus (House mouse)
Collected and cropped from Life (Basel) by CiteAb, provided under a CC-BY license
Image 1 of 41
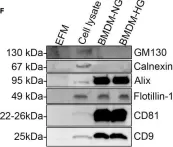
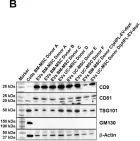
In iScience on 20 August 2021 by Bouchareychas, L., Duong, P., et al.
Fig.1.F

-
WB
-
Collected and cropped from iScience by CiteAb, provided under a CC-BY license
Image 1 of 41
In PLoS Biol on 1 April 2021 by Peruzzotti-Jametti, L., Bernstock, J. D., et al.
Fig.2.A

-
WB
-
Collected and cropped from PLoS Biol by CiteAb, provided under a CC-BY license
Image 1 of 41
In Elife on 9 December 2020 by Vergarajauregui, S., Becker, R., et al.
Fig.3.A

-
ICC-IF
-
Collected and cropped from Elife by CiteAb, provided under a CC-BY license
Image 1 of 41
In Elife on 9 December 2020 by Vergarajauregui, S., Becker, R., et al.
Fig.3.F

-
ICC-IF
-
Collected and cropped from Elife by CiteAb, provided under a CC-BY license
Image 1 of 41
In MBio on 31 March 2020 by Smith, E. P., Cotto-Rosario, A., et al.
Fig.4.A

-
ICC-IF
-
Collected and cropped from MBio by CiteAb, provided under a CC-BY license
Image 1 of 41
In Int J Mol Sci on 17 December 2019 by Higa, N., Shinsato, Y., et al.
Fig.3.D

-
WB
-
Collected and cropped from Int J Mol Sci by CiteAb, provided under a CC-BY license
Image 1 of 41
In Int J Mol Sci on 17 December 2019 by Higa, N., Shinsato, Y., et al.
Fig.3.E

-
WB
-
Collected and cropped from Int J Mol Sci by CiteAb, provided under a CC-BY license
Image 1 of 41
In PLoS One on 12 April 2019 by Duong, P., Chung, A., et al.
Fig.4.A

-
WB
-
Collected and cropped from PLoS One by CiteAb, provided under a CC-BY license
Image 1 of 41
In PLoS One on 8 March 2019 by Lu, T., Zou, Y., et al.
Fig.6.A

-
ICC-IF
-
Homo sapiens (Human)
Collected and cropped from PLoS One by CiteAb, provided under a CC-BY license
Image 1 of 41
In Cell Commun Signal on 27 April 2018 by Hsu, R. M., Zhong, C. Y., et al.
Fig.5.B

-
WB
-
Homo sapiens (Human)
Collected and cropped from Cell Commun Signal by CiteAb, provided under a CC-BY license
Image 1 of 41
In Nat Commun on 23 January 2018 by Kathayat, R. S., Cao, Y., et al.
Fig.7.C

-
WB
-
Collected and cropped from Nat Commun by CiteAb, provided under a CC-BY license
Image 1 of 41
In Int J Mol Sci on 1 July 2017 by Pachler, K., Ketterl, N., et al.
Fig.1.B

-
WB
-
Homo sapiens (Human)
Collected and cropped from Int J Mol Sci by CiteAb, provided under a CC-BY license
Image 1 of 41