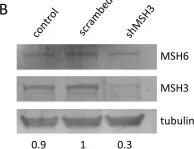
Expansion mutations of triplet repeat sequences cause numerous inherited neurological diseases. In some diseases, affected individuals display somatic expansions in affected tissues which have been linked to accelerated disease onset and progression. There is currently considerable interest in developing therapies to slow somatic repeat expansions to delay or block disease onset. In vitro assays are particularly important to evaluate potential therapeutic interventions. Current assays typically use physical methods to monitor triplet repeat lengths within a population of cells. While useful, most of these assays are relatively slow (∼six weeks) and are somewhat limited in sensitivity to rare events. Here, a new assay, called TRX, is described to monitor CAG•CTG triplet repeat expansions more rapidly and with better sensitivity. TRX uses human tissue culture cells expressing two fluorescent proteins. Red fluorescent protein TagRFP658 is constitutively expressed and serves as an internal control. GFP is expressed in a CAG•CTG repeat length-dependent manner, with longer repeat lengths predicted to give higher green fluorescence intensity. Standard flow cytometry allows quantification of changes in fluorescent signal as a simple readout with <2% sensitivity. Two independently derived cell lines with 63 or 59 CAG repeats yielded similar rates of TRX activity. Cells with increased green fluorescence were observed within one to two weeks of culture, with longer times leading to additional signal. The appearance of green fluorescence was partly dependent on MutSβ, the DNA MSH2-MSH3 complex, based on siRNA knockdown of MSH3. However, physical analysis of the CAG•CTG repeat tracts by MiSeq deep sequencing or capillary electrophoresis showed limited changes in the length of the repeat tracts. We conclude that the TRX assay is a promising new tool for monitoring CAG•CTG repeat expansions but that further development of the assay is needed to make it fully useful.
Product Citations: 95
The TRX assay for triplet repeat expansions
Preprint on BioRxiv : the Preprint Server for Biology on 20 December 2024 by O’Brien, T., Salciute, K., et al.
-
FC/FACS
In Nature Communications on 25 September 2023 by Omatsu, M., Nakanishi, Y., et al.
Mesenchymal activation, characterized by dense stromal infiltration of immune and mesenchymal cells, fuels the aggressiveness of colorectal cancers (CRC), driving progression and metastasis. Targetable molecules in the tumor microenvironment (TME) need to be identified to improve the outcome in CRC patients with this aggressive phenotype. This study reports a positive link between high thrombospondin-1 (THBS1) expression and mesenchymal characteristics, immunosuppression, and unfavorable CRC prognosis. Bone marrow-derived monocyte-like cells recruited by CXCL12 are the primary source of THBS1, which contributes to the development of metastasis by inducing cytotoxic T-cell exhaustion and impairing vascularization. Furthermore, in orthotopically generated CRC models in male mice, THBS1 loss in the TME renders tumors partially sensitive to immune checkpoint inhibitors and anti-cancer drugs. Our study establishes THBS1 as a potential biomarker for identifying mesenchymal CRC and as a critical suppressor of antitumor immunity that contributes to the progression of this malignancy with a poor prognosis.
© 2023. Springer Nature Limited.
-
IHC
-
Homo sapiens (Human)
-
Cancer Research
In Frontiers in Immunology on 27 January 2023 by Spanjaard, A., Stratigopoulou, M., et al.
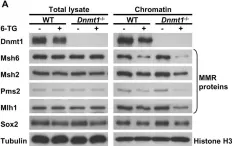
The development and differentiation of B cells is intimately linked to cell proliferation and the generation of diverse immunoglobulin gene (Ig) repertoires. The ubiquitin E3 ligase HUWE1 controls proliferation, DNA damage responses, and DNA repair, including the base excision repair (BER) pathway. These processes are of crucial importance for B-cell development in the bone marrow, and the germinal center (GC) response, which results in the clonal expansion and differentiation of B cells expressing high affinity immunoglobulins. Here, we re-examined the role of HUWE1 in B-cell proliferation and Ig gene diversification, focusing on its involvement in somatic hypermutation (SHM) and class switch recombination (CSR). B-cell-specific deletion of Huwe1 resulted in impaired development, differentiation and maturation of B cells in the bone marrow and peripheral lymphoid organs. HUWE1 deficiency diminished SHM and CSR by impairing B-cell proliferation and AID expression upon activation in vitro and in vivo, and was unrelated to the HUWE1-dependent regulation of the BER pathway. Interestingly, we found that HUWE1-deficient B cells showed increased mRNA expression of Myc target genes upon in vitro activation despite diminished proliferation. Our results confirm that the E3 ligase HUWE1 is an important contributor in coordinating the rapid transition of antigen naïve, resting B cells into antigen-activated B cells and regulates mutagenic processes in B cells by controlling AID expression and the post-transcriptional output of Myc target genes.
Copyright © 2023 Spanjaard, Stratigopoulou, de Groot, Aslam, van den Berk, Stappenbelt, Ayidah, Catsman, Pardieck, Kreft, Arens, Guikema and Jacobs.
-
WB
-
Immunology and Microbiology
In American Journal of Cancer Research on 12 January 2023 by Mai, R. T., Chao, C. H., et al.
Numerous reports indicate that enhanced expression of Y-box binding protein-1 (YB-1) in tumor cells is strongly associated with tumorigenesis, aggressiveness, drug resistance, as well as poor prognosis in several types of cancers, and YB-1 is considered to be an oncogene. The molecular mechanism contributing to the regulation of the biological activities of YB-1 remains obscure. Sumoylation, a post-translational modification involving the covalent conjugation of small ubiquitin-like modifier (SUMO) proteins to a target protein, plays key roles in the modulation of protein functions. In this study, our results revealed that YB-1 is sumoylated and that Lys26 is a critical residue for YB-1 sumoylation. Moreover, YB-1 was found to directly interact with SUMO proteins, and disruption of the SUMO-interacting motif (SIM) of YB-1 not only interfered with this interaction but also diminished YB-1 sumoylation. The subcellular localization, protein stability, and transcriptional regulatory activity of YB-1 were not significantly affected by sumoylation. However, decreased sumoylation disrupted the interaction between YB-1 and PCNA as well as YB-1-mediated inhibition of the MutSα/PCNA interaction and MutSα mismatch binding activity, indicating a functional role of YB-1 sumoylation in inducing DNA mismatch repair (MMR) deficiency and spontaneous mutations. The MMR machinery also recognizes alkylator-modified DNA adducts to signal for cell death. We further demonstrated that YB-1 sumoylation is crucial for the inhibition of SN1-type alkylator MNNG-induced cytotoxicity, G2/M-phase arrest, apoptosis, and the MMR-dependent DNA damage response. Collectively, these results provide molecular explanations for the impact of YB-1 sumoylation on MMR deficiency and alkylator tolerance, which may provide insight for designing therapeutic strategies for malignancies and alkylator-resistant cancers associated with YB-1 overexpression.
AJCR Copyright © 2022.
-
Cancer Research
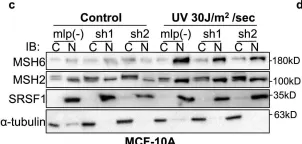
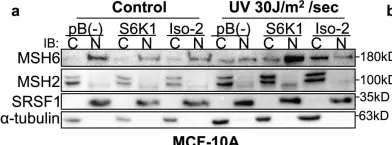
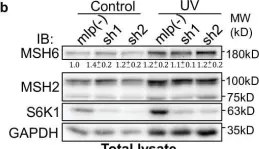
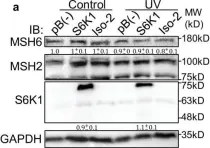
S6K1 phosphorylates Cdk1 and MSH6 to regulate DNA repair.
In eLife on 3 October 2022 by Amar-Schwartz, A., Ben Hur, V., et al.
The mTORC1 substrate, S6 Kinase 1 (S6K1), is involved in the regulation of cell growth, ribosome biogenesis, glucose homeostasis, and adipogenesis. Accumulating evidence has suggested a role for mTORC1 signaling in the DNA damage response. This is mostly based on the findings that mTORC1 inhibitors sensitized cells to DNA damage. However, a direct role of the mTORC1-S6K1 signaling pathway in DNA repair and the mechanism by which this signaling pathway regulates DNA repair is unknown. In this study, we discovered a novel role for S6K1 in regulating DNA repair through the coordinated regulation of the cell cycle, homologous recombination (HR) DNA repair (HRR) and mismatch DNA repair (MMR) mechanisms. Here, we show that S6K1 orchestrates DNA repair by phosphorylation of Cdk1 at serine 39, causing G2/M cell cycle arrest enabling homologous recombination and by phosphorylation of MSH6 at serine 309, enhancing MMR. Moreover, breast cancer cells harboring RPS6KB1 gene amplification show increased resistance to several DNA damaging agents and S6K1 expression is associated with poor survival of breast cancer patients treated with chemotherapy. Our findings reveal an unexpected function of S6K1 in the DNA repair pathway, serving as a tumorigenic barrier by safeguarding genomic stability.
© 2022, Amar-Schwartz, Ben Hur, Jbara et al.
-
WB
-
Genetics
In Front Immunol on 27 January 2023 by Spanjaard, A., Stratigopoulou, M., et al.
Fig.4.B

-
WB
-
Collected and cropped from Front Immunol by CiteAb, provided under a CC-BY license
Image 1 of 11
In Elife on 3 October 2022 by Amar-Schwartz, A., Ben Hur, V., et al.
Fig.4.C

-
WB
-
Collected and cropped from Elife by CiteAb, provided under a CC-BY license
Image 1 of 11
In Elife on 3 October 2022 by Amar-Schwartz, A., Ben Hur, V., et al.
Fig.4.A

-
WB
-
Collected and cropped from Elife by CiteAb, provided under a CC-BY license
Image 1 of 11
In Elife on 3 October 2022 by Amar-Schwartz, A., Ben Hur, V., et al.
Fig.4.B

-
WB
-
Collected and cropped from Elife by CiteAb, provided under a CC-BY license
Image 1 of 11
In Elife on 3 October 2022 by Amar-Schwartz, A., Ben Hur, V., et al.
Fig.4.A

-
WB
-
Collected and cropped from Elife by CiteAb, provided under a CC-BY license
Image 1 of 11
In Sci Rep on 25 November 2016 by Wang, K. Y., Chen, C. C., et al.
Fig.4.A

-
WB
-
Collected and cropped from Sci Rep by CiteAb, provided under a CC-BY license
Image 1 of 11
In Sci Rep on 5 June 2015 by Nakatani, R., Nakamori, M., et al.
Fig.5.B

-
WB
-
Collected and cropped from Sci Rep by CiteAb, provided under a CC-BY license
Image 1 of 11
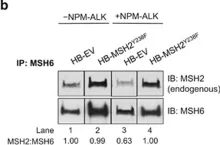
In Blood Cancer J on 15 May 2015 by Bone, K. M., Wang, P., et al.
Fig.3.A

-
WB
-
Homo sapiens (Human)
Collected and cropped from Blood Cancer J by CiteAb, provided under a CC-BY license
Image 1 of 11
In Blood Cancer J on 15 May 2015 by Bone, K. M., Wang, P., et al.
Fig.3.B

-
WB
-
Homo sapiens (Human)
Collected and cropped from Blood Cancer J by CiteAb, provided under a CC-BY license
Image 1 of 11
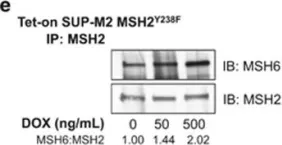
In Blood Cancer J on 15 May 2015 by Bone, K. M., Wang, P., et al.
Fig.4.E

-
WB
-
Homo sapiens (Human)
Collected and cropped from Blood Cancer J by CiteAb, provided under a CC-BY license
Image 1 of 11
In PLoS One on 5 December 2012 by Campregher, C., Schmid, G., et al.
Fig.2.B

-
WB
-
Collected and cropped from PLoS One by CiteAb, provided under a CC-BY license
Image 1 of 11