Localization to the endoplasmic reticulum (ER) and subsequent disulfide bond formation are crucial processes governing the biogenesis of secretory pathway proteins in eukaryotes. Hence, comprehending the mechanisms underlying these processes is important. Here, we have engineered firefly luciferase (FLuc) as a tool to detect deficiencies in these processes within mammalian cells. To achieve this, we introduced multiple cysteine substitutions into FLuc and targeted it to the ER. The reporter exhibited FLuc activity in response to defects in protein localization or disulfide bond formation within the ER. Notably, this system exhibited outstanding sensitivity, reproducibility, and convenience in detecting abnormalities in these processes. We applied this system to observe a protein translocation defect induced by an inhibitor of HIV receptor biogenesis. Moreover, utilizing the system, we showed that modulating LMF1 levels dramatically impacted the ER's redox environment, confirming that LMF1 plays some critical role in the redox control of the ER.
© 2024 The Author(s).
Product Citations: 15
In IScience on 15 November 2024 by Kadokura, H., Harada, N., et al.
The nucleolar phase of signal recognition particle assembly.
In Life Science Alliance on 1 August 2024 by Issa, A., Schlotter, F., et al.
The signal recognition particle is essential for targeting transmembrane and secreted proteins to the endoplasmic reticulum. Remarkably, because they work together in the cytoplasm, the SRP and ribosomes are assembled in the same biomolecular condensate: the nucleolus. How important is the nucleolus for SRP assembly is not known. Using quantitative proteomics, we have investigated the interactomes of SRP components. We reveal that SRP proteins are associated with scores of nucleolar proteins important for ribosome biogenesis and nucleolar structure. Having monitored the subcellular distribution of SRP proteins upon controlled nucleolar disruption, we conclude that an intact organelle is required for their proper localization. Lastly, we have detected two SRP proteins in Cajal bodies, which indicates that previously undocumented steps of SRP assembly may occur in these bodies. This work highlights the importance of a structurally and functionally intact nucleolus for efficient SRP production and suggests that the biogenesis of SRP and ribosomes may be coordinated in the nucleolus by common assembly factors.
© 2024 Issa et al.
-
Homo sapiens (Human)
In Journal of Molecular Biology on 15 March 2024 by Miller, S. C., Tikhonova, E. B., et al.
Many insulin gene variants alter the protein sequence and result in monogenic diabetes due to insulin insufficiency. However, the molecular mechanisms of various disease-causing mutations are unknown. Insulin is synthesized as preproinsulin containing a signal peptide (SP). SPs of secreted proteins are recognized by the signal recognition particle (SRP) or by another factor in a SRP-independent pathway. If preproinsulin uses SRP-dependent or independent pathways is still debatable. We demonstrate by the use of site-specific photocrosslinking that the SRP subunit, SRP54, interacts with the preproinsulin SP. Moreover, SRP54 depletion leads to the decrease of insulin mRNA and protein expression, supporting the involvement of the RAPP protein quality control in insulin biogenesis. RAPP regulates the quality of secretory proteins through degradation of their mRNA. We tested five disease-causing mutations in the preproinsulin SP on recognition by SRP and on their effects on mRNA and protein levels. We demonstrate that the effects of mutations are associated with their position in the SP and their severity. The data support diverse molecular mechanisms involved in the pathogenesis of these mutations. We show for the first time the involvement of the RAPP protein quality control pathway in insulin biogenesis that is implicated in the development of neonatal diabetes caused by the Leu13Arg mutation.
Copyright © 2024 Elsevier Ltd. All rights reserved.
-
WB
-
Biochemistry and Molecular biology
-
Genetics
SRP19and the protein secretion machinery is a targetable vulnerability in cancers withAPCloss
Preprint on BioRxiv : the Preprint Server for Biology on 31 January 2024 by Xi, X., Liu, L., et al.
Adenomatous Polyposis Coli ( APC ) is a tumour suppressor that is frequently lost in colorectal and other cancers. A common mechanism for APC loss includes heterozygous APC deletion. Here, we show that SRP19 , is located near APC and is often co-deleted in these tumours. Heterozygous APC/SRP19 loss leads to lower levels of SRP19 mRNA and protein. Consequently, cells with APC/SRP19 loss are vulnerable to partial suppression of SRP19 . We show that SRP19 loss is a unique vulnerability since SRP19 is rate limiting for the formation of the Signal Recognition Particle (SRP), a complex that mediates translocation of proteins to the ER. Consistent with these observations, partial SRP19 knock-down or low dose Arsenic Trioxide treatment induces ER stress and inhibits proliferation in APC/SRP19 loss cancers. Our work identifies a new strategy to treat cancers with APC/SRP19 heterozygous deletions and provides a framework for identifying vulnerabilities associated with loss of a tumour suppressor.
Mechanism of signal-anchor triage during early steps of membrane protein insertion.
In Molecular Cell on 16 March 2023 by Wu, H. & Hegde, R. S.
Most membrane proteins use their first transmembrane domain, known as a signal anchor (SA), for co-translational targeting to the endoplasmic reticulum (ER) via the signal recognition particle (SRP). The SA then inserts into the membrane using either the Sec61 translocation channel or the ER membrane protein complex (EMC) insertase. How EMC and Sec61 collaborate to ensure SA insertion in the correct topology is not understood. Using site-specific crosslinking, we detect a pre-insertion SA intermediate adjacent to EMC. This intermediate forms after SA release from SRP but before ribosome transfer to Sec61. The polypeptide's N-terminal tail samples a cytosolic vestibule bordered by EMC3, from where it can translocate across the membrane concomitant with SA insertion. The ribosome then docks on Sec61, which has an opportunity to insert those SAs skipped by EMC. These results suggest that EMC acts between SRP and Sec61 to triage SAs for insertion during membrane protein biogenesis.
Copyright © 2023 MRC Laboratory of Molecular Biology. Published by Elsevier Inc. All rights reserved.
-
Biochemistry and Molecular biology
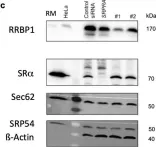
In Molecules on 11 June 2021 by Bhadra, P., Schorr, S., et al.
Fig.4.C

-
WB
-
Collected and cropped from Molecules by CiteAb, provided under a CC-BY license
Image 1 of 1