The DEK chromatin remodeling protein has oncogenic functions in breast cancers, but its functional role in normal mammary gland epithelium has remained unexplored. We developed two novel genetically engineered mouse models to study the role of Dek in normal mammary gland biology in vivo. Mammary gland-specific Dek transgenic mice developed hyperplasia and had a transcriptional profile that revealed increased expression of cell cycle, mammary stem/progenitor, and lactation-associated genes. Conversely, Dek knockout mice exhibited mammary gland functional defects resulting in dramatically reduced pup survival. Analysis of previously published scRNA-sequencing of mouse mammary glands revealed that Dek is most highly expressed in mammary stem cells and alveolar progenitor cells, supporting the observed phenotypes. Mechanistically, we discovered that Dek is a modifier of Ezh2 methyltransferase activity, up-regulating the levels of histone H3K27me3 to control gene transcription. Combined, this is the first report to show that Dek promotes proliferation of mammary epithelial cells via transcriptional deregulation of cell cycle genes, potentially via epigenetic mechanisms, in vivo.
© 2025 Johnstone et al.
Product Citations: 31
DEK promotes mammary hyperplasia and is associated with H3K27me3 epigenetic modifications.
In Life Science Alliance on 1 September 2025 by Johnstone, M. E., Leck, A. L., et al.
-
Genetics
Structural insights into how DEK nucleosome binding facilitates H3K27 trimethylation in chromatin.
In Nature Structural Molecular Biology on 21 February 2025 by Kujirai, T., Echigoya, K., et al.
Structural diversity of the nucleosome affects chromatin conformations and regulates eukaryotic genome functions. Here we identify DEK, whose function is unknown, as a nucleosome-binding protein. In embryonic neural progenitor cells, DEK colocalizes with H3 K27 trimethylation (H3K27me3), the facultative heterochromatin mark. DEK stimulates the methyltransferase activity of Polycomb repressive complex 2 (PRC2), which is responsible for H3K27me3 deposition in vitro. Cryo-electron microscopy structures of the DEK-nucleosome complexes reveal that DEK binds the nucleosome by its tripartite DNA-binding mode on the dyad and linker DNAs and interacts with the nucleosomal acidic patch by its newly identified histone-binding region. The DEK-nucleosome interaction mediates linker DNA reorientation and induces chromatin compaction, which may facilitate PRC2 activation. These findings provide mechanistic insights into chromatin structure-mediated gene regulation by DEK.
© 2025. The Author(s).
-
ChIP
-
Biochemistry and Molecular biology
-
Genetics
In Blood Cancer Journal on 9 October 2024 by Hopper, M. A., Dropik, A. R., et al.
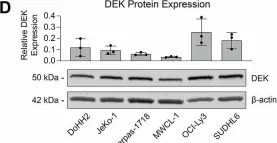
This study sheds light on the pivotal role of the oncoprotein DEK in B-cell lymphoma. We reveal DEK expression correlates with increased tumor proliferation and inferior overall survival in cases diagnosed with low-grade B-cell lymphoma (LGBCL). We also found significant correlation between DEK expression and copy number alterations in LGBCL tumors, highlighting a novel mechanism of LGBCL pathogenesis that warrants additional exploration. To interrogate the mechanistic role of DEK in B-cell lymphoma, we generated a DEK knockout cell line model, which demonstrated DEK depletion caused reduced proliferation and altered expression of key cell cycle and apoptosis-related proteins, including Bcl-2, Bcl-xL, and p53. Notably, DEK depleted cells showed increased sensitivity to apoptosis-inducing agents, including venetoclax and staurosporine, which underscores the therapeutic potential of targeting DEK in B-cell lymphomas. Overall, our study contributes to a better understanding of DEK's role as an oncoprotein in B-cell lymphomas, highlighting its potential as both a promising therapeutic target and a novel biomarker for aggressive LGBCL. Further research elucidating the molecular mechanisms underlying DEK-mediated tumorigenesis could pave the way for improved treatment strategies and better clinical outcomes for patients with B-cell lymphoma.
© 2024. The Author(s).
-
ICC-IF
-
WB
-
Cancer Research
-
Immunology and Microbiology
Preprint on BioRxiv : the Preprint Server for Biology on 10 September 2024 by Johnstone, M., Leck, A., et al.
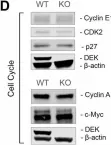
The DEK chromatin remodeling protein was previously shown to confer oncogenic phenotypes to human and mouse mammary epithelial cells using in vitro and knockout mouse models. However, its functional role in normal mammary gland epithelium remained unexplored. We developed two novel mouse models to study the role of Dek in normal mammary gland biology in vivo . Mammary gland-specific Dek over-expression in mice resulted in hyperproliferation of cells that visually resembled alveolar cells, and a transcriptional profile that indicated increased expression of cell cycle, mammary stem/progenitor, and lactation-associated genes. Conversely, Dek knockout mice exhibited an alveologenesis or lactation defect, resulting in dramatically reduced pup survival. Analysis of previously published single-cell RNA-sequencing of mouse mammary glands revealed that Dek is most highly expressed in mammary stem cells and alveolar progenitor cells, and to a lesser extent in basal epithelial cells, supporting the observed phenotypes. Mechanistically, we discovered that Dek is a modifier of Ezh2 methyltransferase activity, upregulating the levels of histone H3 trimethylation on lysine 27 (H3K27me3) to control gene transcription. Combined, this work indicates that Dek promotes proliferation of mammary epithelial cells via cell cycle deregulation. Furthermore, we report a novel function for Dek in alveologenesis and histone H3 K27 trimethylation.
-
Genetics
In The Journal of Biological Chemistry on 1 December 2023 by Chen, X., Pillay, S., et al.
EKLF/KLF1 is an essential transcription factor that plays a global role in erythroid transcriptional activation. Regulation of KLF1 is of interest, as it displays a highly restricted expression pattern, limited to erythroid cells and its progenitors. Here we use biochemical affinity purification to identify the DDX5/p68 protein as an activator of KLF1 by virtue of its interaction with the erythroid-specific DNAse hypersensitive site upstream enhancer element (EHS1). We further show that this protein associates with DEK and CTCF. We postulate that the range of interactions of DDX5/p68 with these and other proteins known to interact with this element render it part of the enhanseosome complex critical for optimal expression of KLF1 and enables the formation of a proper chromatin configuration at the Klf1 locus. These individual interactions provide quantitative contributions that, in sum, establish the high-level activity of the Klf1 promoter and suggest they can be selectively manipulated for clinical benefit.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
-
Biochemistry and Molecular biology
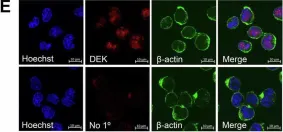
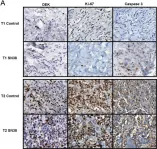
In Blood Cancer J on 9 October 2024 by Hopper, M. A., Dropik, A. R., et al.
Fig.2.E

-
ICC-IF
-
Collected and cropped from Blood Cancer J by CiteAb, provided under a CC-BY license
Image 1 of 9
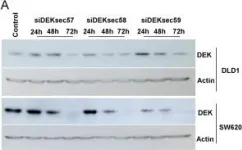
In Blood Cancer J on 9 October 2024 by Hopper, M. A., Dropik, A. R., et al.
Fig.2.D

-
WB
-
Collected and cropped from Blood Cancer J by CiteAb, provided under a CC-BY license
Image 1 of 9
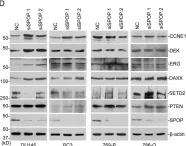
In Blood Cancer J on 9 October 2024 by Hopper, M. A., Dropik, A. R., et al.
Fig.3.D

-
WB
-
Collected and cropped from Blood Cancer J by CiteAb, provided under a CC-BY license
Image 1 of 9
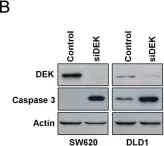
In Cell Death Differ on 1 June 2019 by Ju, L. G., Zhu, Y., et al.
Fig.6.D

-
WB
-
Homo sapiens (Human)
Collected and cropped from Cell Death Differ by CiteAb, provided under a CC-BY license
Image 1 of 9
In Cell Death Differ on 1 June 2019 by Ju, L. G., Zhu, Y., et al.
Fig.6.E

-
WB
-
Homo sapiens (Human)
Collected and cropped from Cell Death Differ by CiteAb, provided under a CC-BY license
Image 1 of 9
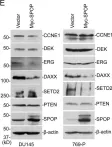
In BMC Cancer on 16 December 2014 by Martinez-Useros, J., Rodriguez-Remirez, M., et al.
Fig.4.B

-
WB
-
Homo sapiens (Human)
Collected and cropped from BMC Cancer by CiteAb, provided under a CC-BY license
Image 1 of 9
In BMC Cancer on 16 December 2014 by Martinez-Useros, J., Rodriguez-Remirez, M., et al.
Fig.2.A

-
WB
-
Collected and cropped from BMC Cancer by CiteAb, provided under a CC-BY license
Image 1 of 9
In BMC Cancer on 16 December 2014 by Martinez-Useros, J., Rodriguez-Remirez, M., et al.
Fig.4.A

-
IHC
-
Collected and cropped from BMC Cancer by CiteAb, provided under a CC-BY license
Image 1 of 9
In PLoS Genet on 1 September 2014 by Iannetti, A., Ledoux, A. C., et al.
Fig.5.E

-
WB
-
Collected and cropped from PLoS Genet by CiteAb, provided under a CC-BY license
Image 1 of 9