Cell senescence impedes the self‑renewal and osteogenic capacity of bone marrow mesenchymal stem cells (BMSCs), thus limiting their application in tissue regeneration. The present study aimed to elucidate the role and mechanism of repetitive element (RE) activation in BMSC senescence and osteogenesis, as well as the intervention effect of quercetin. In an H2O2‑induced BMSC senescence model, quercetin treatment alleviated senescence as shown by a decrease in senescence‑associated β‑galactosidase (SA‑β‑gal)‑positive cell ratio, increased colony formation ability and decreased mRNA expression of p21 and senescence‑associated secretory phenotype genes. DNA damage response marker γ‑H2AX increased in senescent BMSCs, while expression of epigenetic markers methylation histone H3 Lys9, heterochromatin protein 1α and heterochromatin‑related nuclear membrane protein lamina‑associated polypeptide 2 decreased. Quercetin rescued these alterations, indicating its ability to ameliorate senescence by stabilizing heterochromatin structure where REs are primarily suppressed. Transcriptional activation of REs accompanied by accumulation of cytoplasmic double‑stranded (ds)RNA, as well as triggering of the RNA sensor retinoic acid‑inducible gene I (RIG‑I) receptor pathway in H2O2‑induced senescent BMSCs were shown. Similarly, quercetin treatment inhibited these responses. Additionally, RIG‑I knockdown led to a decreased number of SA‑β‑gal‑positive cells, confirming its functional impact on senescence. Induction of senescence or administration of dsRNA analogue significantly hindered the osteogenic capacity of BMSCs, while quercetin treatment or RIG‑I knockdown reversed the decline in osteogenic function. The findings of the current study demonstrated that quercetin inhibited the activation of REs and the RIG‑I RNA sensing pathway via epigenetic regulation, thereby alleviating the senescence of BMSCs and promoting osteogenesis.
Product Citations: 69
In International Journal of Molecular Medicine on 1 January 2025 by Sun, Y., Wang, C., et al.
-
Genetics
Preprint on BioRxiv : the Preprint Server for Biology on 23 December 2024 by Lewis, R., Sinigiani, V., et al.
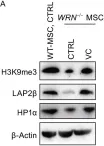
Summary In most eukaryotic cells, euchromatin is localized in the nuclear interior, whereas heterochromatin is enriched at the nuclear envelope (NE). This conventional chromatin organization is established by heterochromatin tethering to the NE, however its importance for cellular homeostasis is largely unexplored. Peripheral heterochromatin localization relies on redundant NE-tethering systems. One tether is constituted by the lamin B receptor (LBR) in mammals, but the enigmatic nature of the other tethers has hampered functional analyses. Here we demonstrate that the downregulation of abundant, ubiquitous NE proteins can induce the global detachment of heterochromatin from the NE. Among these factors, we identify LBR and LAP2 as major players in bulk heterochromatin attachment to the NE in pluripotent and differentiated mammalian cells. Their loss leads to repositioning of heterochromatin to the nuclear interior, changes in chromatin accessibility, deregulation of gene expression including activation of antiviral innate immunity, and defects in cell fate determination.
In Nucleus on 1 December 2024 by Wu, Z., Omura, I., et al.
The Nuclear envelope (NE) is frequently challenged by mechanical stimuli involving cells passing through a tight space and such stress is known as "NE stress." Various factors that cooperate to repair the NE have been identified, including endosomal sorting complex required for transport-III (ESCRT-III). Recently, vacuolar protein sorting 4 homolog B (VPS4B) has been reported to modulate the recycling of ESCRT-III during NE repair, but the regulatory mechanism remains unclear. Our previous study revealed that U251MG cells, derived from the glioblastoma (GBM), exhibited nuclear deformation followed by DNA damage upon mechanical NE stress while these phenotypes were not observed in U87MG, another GBM-derived cell line. Here, we found that VPS4B expression was lower in U251MG than in U87MG. Our functional analysis demonstrated that insufficient VPS4B triggers an inadequate response to NE stress and that VPS4B regulates the dynamics of ESCRT-III, uncovering the mechanism underlying the NE stress response in GBM.
CRISPR screening uncovers nucleolar RPL22 as a heterochromatin destabilizer and senescence driver.
In Nucleic Acids Research on 28 October 2024 by Li, H. Y., Wang, M., et al.
Dysfunction of the ribosome manifests during cellular senescence and contributes to tissue aging, functional decline, and development of aging-related disorders in ways that have remained enigmatic. Here, we conducted a comprehensive CRISPR-based loss-of-function (LOF) screen of ribosome-associated genes (RAGs) in human mesenchymal progenitor cells (hMPCs). Through this approach, we identified ribosomal protein L22 (RPL22) as the foremost RAG whose deficiency mitigates the effects of cellular senescence. Consequently, absence of RPL22 delays hMPCs from becoming senescent, while an excess of RPL22 accelerates the senescence process. Mechanistically, we found in senescent hMPCs, RPL22 accumulates within the nucleolus. This accumulation triggers a cascade of events, including heterochromatin decompaction with concomitant degradation of key heterochromatin proteins, specifically heterochromatin protein 1γ (HP1γ) and heterochromatin protein KRAB-associated protein 1 (KAP1). Subsequently, RPL22-dependent breakdown of heterochromatin stimulates the transcription of ribosomal RNAs (rRNAs), triggering cellular senescence. In summary, our findings unveil a novel role for nucleolar RPL22 as a destabilizer of heterochromatin and a driver of cellular senescence, shedding new light on the intricate mechanisms underlying the aging process.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
-
WB
-
ICC-IF
-
Biochemistry and Molecular biology
In Cell Stem Cell on 1 August 2024 by Saurat, N., Minotti, A. P., et al.
Aging is the biggest risk factor for the development of Alzheimer's disease (AD). Here, we performed a whole-genome CRISPR screen to identify regulators of neuronal age and show that the neddylation pathway regulates both cellular age and AD neurodegeneration in a human stem cell model. Specifically, we demonstrate that blocking neddylation increased cellular hallmarks of aging and led to an increase in Tau aggregation and phosphorylation in neurons carrying the APPswe/swe mutation. Aged APPswe/swe but not isogenic control neurons also showed a progressive decrease in viability. Selective neuronal loss upon neddylation inhibition was similarly observed in other isogenic AD and in Parkinson's disease (PD) models, including PSENM146V/M146V cortical and LRRK2G2019S/G2019S midbrain dopamine neurons, respectively. This study indicates that cellular aging can reveal late-onset disease phenotypes, identifies new potential targets to modulate AD progression, and describes a strategy to program age-associated phenotypes into stem cell models of disease.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
-
Neuroscience
-
Stem Cells and Developmental Biology
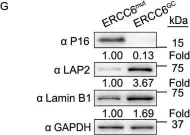
In Protein Cell on 1 January 2020 by Wang, S., Min, Z., et al.
Fig.3.G

-
WB
-
Collected and cropped from Protein Cell by CiteAb, provided under a CC-BY license
Image 1 of 6
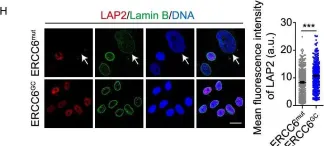
In Protein Cell on 1 January 2020 by Wang, S., Min, Z., et al.
Fig.3.H

-
ICC-IF
-
Collected and cropped from Protein Cell by CiteAb, provided under a CC-BY license
Image 1 of 6
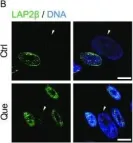
In Protein Cell on 1 June 2019 by Geng, L., Liu, Z., et al.
Fig.5.B

-
ICC-IF
-
Homo sapiens (Human)
Collected and cropped from Protein Cell by CiteAb, provided under a CC-BY license
Image 1 of 6
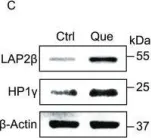
In Protein Cell on 1 June 2019 by Geng, L., Liu, Z., et al.
Fig.5.C

-
WB
-
Homo sapiens (Human)
Collected and cropped from Protein Cell by CiteAb, provided under a CC-BY license
Image 1 of 6
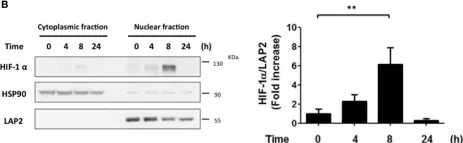
In Front Cell Infect Microbiol on 13 April 2017 by Canales, J., Valenzuela, M., et al.
Fig.1.B

-
WB
-
Collected and cropped from Front Cell Infect Microbiol by CiteAb, provided under a CC-BY license
Image 1 of 6
In Protein Cell on 1 July 2016 by Li, Y., Zhang, W., et al.
Fig.3.A

-
WB
-
Mus musculus (House mouse)
Collected and cropped from Protein Cell by CiteAb, provided under a CC-BY license
Image 1 of 6