A critical yet challenging step in protein complex assembly is the formation of a dimeric intermediate that serves as a seed for incorporating additional subunits. We hypothesized that this step could be facilitated by "bi-handed" chaperones that recognize two different subunits through distinct domains (hands). However, whether such chaperones exist remained unknown. Here, we identify AAGAB as a bona fide bi-handed chaperone. AAGAB uses its C-terminal domain (CTD) to bind the α subunit and its GTPase-like domain (GD) to bind the σ2 subunit of the AP2 adaptor complex, a central player in membrane trafficking. AAGAB first recruits α via its CTD; σ2 then joins through interaction with α, forming a conformationally immature α:σ2 hemicomplex at the CTD. This hemicomplex is subsequently transferred to the GD via a GD:σ2 binding interface, accompanied by conformational maturation. These findings establish AAGAB as the founding member of a bi-handed chaperone family and reveal an intramolecular handover mechanism that underlies their mode of action.
Product Citations: 32
In Science Advances on 12 September 2025 by Wu, J., Wan, C., et al.
Preprint on BioRxiv : the Preprint Server for Biology on 16 February 2025 by Tempes, A., Brzozowska, A., et al.
Clathrin-mediated endocytosis (CME) is a process in which ligands and their corresponding receptors at the cell surface are internalized via clathrin-coated invaginations of the plasma membrane. Both clathrin and endocytic cargo are recruited to the clathrin-coated pit by the adaptor protein complex AP2. AP2 resides in the cytoplasm in the closed conformation and opens upon interacting with the plasma membrane. Effective CME requires both pools of AP2, open and closed, and effective transition between these states but the mechanisms regulating this transition are only partially understood. Here, we report that serine 45 (S45) of the µ2 subunit of the AP2 complex is phosphorylated in a p70S6 kinase-dependent manner in HeLa cells both with hyperactivated mTOR signaling and under basal conditions. We demonstrate that the loss of S45-µ2 phosphorylation results in decreased internalization of canonical CME cargo such as transferrin and PDGF receptors. In Caenorhabditis elegans , the absence of S45-µ2 phosphorylation produces the dumpy phenotype characteristic of the loss of function of AP2. Our live imaging experiments further suggest that these phenotypes might arise because S45-μ2 phosphorylation is needed for conformational changes of the AP2 complex. These findings uncover a mechanism central to CME control and extend our knowledge on the role of post-translational modifications of AP2 components in regulating the function of this complex.
Rapid turnover of CTLA4 is associated with a complex architecture of reversible ubiquitylation.
In The Journal of Cell Biology on 6 January 2025 by Tey, P. Y., Dufner, A., et al.
The immune checkpoint regulator CTLA4 is an unusually short-lived membrane protein. Here, we show that its lysosomal degradation is dependent on ubiquitylation at lysine residues 203 and 213. Inhibition of the v-ATPase partially restores CTLA4 levels following cycloheximide treatment, but also reveals a fraction that is secreted in exosomes. The endosomal deubiquitylase, USP8, interacts with CTLA4, and its loss enhances CTLA4 ubiquitylation in cancer cells, mouse CD4+ T cells, and cancer cell-derived exosomes. Depletion of the USP8 adapter protein, HD-PTP, but not ESCRT-0 recapitulates this cellular phenotype but shows distinct properties vis-à-vis exosome incorporation. Re-expression of wild-type USP8, but neither a catalytically inactive nor a localization-compromised ΔMIT domain mutant can rescue delayed degradation of CTLA4 or counteract its accumulation in clustered endosomes. UbiCRest analysis of CTLA4-associated ubiquitin chain linkages identifies a complex mixture of conventional Lys63- and more unusual Lys27- and Lys29-linked polyubiquitin chains that may underly the rapidity of protein turnover.
© 2024 Tey et al.
-
WB
-
Cell Biology
Apache is a neuronal player in autophagy required for retrograde axonal transport of autophagosomes.
In Cellular and Molecular Life Sciences : CMLS on 5 October 2024 by Parisi, B., Esposito, A., et al.
Neurons are dependent on efficient quality control mechanisms to maintain cellular homeostasis and function due to their polarization and long-life span. Autophagy is a lysosomal degradative pathway that provides nutrients during starvation and recycles damaged and/or aged proteins and organelles. In neurons, autophagosomes constitutively form in distal axons and at synapses and are trafficked retrogradely to the cell soma to fuse with lysosomes for cargo degradation. How the neuronal autophagy pathway is organized and controlled remains poorly understood. Several presynaptic endocytic proteins have been shown to regulate both synaptic vesicle recycling and autophagy. Here, by combining electron, fluorescence, and live imaging microscopy with biochemical analysis, we show that the neuron-specific protein APache, a presynaptic AP-2 interactor, functions in neurons as an important player in the autophagy process, regulating the retrograde transport of autophagosomes. We found that APache colocalizes and co-traffics with autophagosomes in primary cortical neurons and that induction of autophagy by mTOR inhibition increases LC3 and APache protein levels at synaptic boutons. APache silencing causes a blockade of autophagic flux preventing the clearance of p62/SQSTM1, leading to a severe accumulation of autophagosomes and amphisomes at synaptic terminals and along neurites due to defective retrograde transport of TrkB-containing signaling amphisomes along the axons. Together, our data identify APache as a regulator of the autophagic cycle, potentially in cooperation with AP-2, and hypothesize that its dysfunctions contribute to the early synaptic impairments in neurodegenerative conditions associated with impaired autophagy.
© 2024. The Author(s).
-
WB
-
Biochemistry and Molecular biology
-
Cell Biology
An AAGAB-to-CCDC32 handover mechanism controls the assembly of the AP2 adaptor complex.
In Proceedings of the National Academy of Sciences of the United States of America on 20 August 2024 by Wan, C., Puscher, H., et al.
Vesicular transport relies on multimeric trafficking complexes to capture cargo and drive vesicle budding and fusion. Faithful assembly of the trafficking complexes is essential to their functions but remains largely unexplored. Assembly of AP2 adaptor, a heterotetrameric protein complex regulating clathrin-mediated endocytosis, is assisted by the chaperone AAGAB. Here, we found that AAGAB initiates AP2 assembly by stabilizing its α and σ2 subunits, but the AAGAB:α:σ2 complex cannot recruit additional AP2 subunits. We identified CCDC32 as another chaperone regulating AP2 assembly. CCDC32 recognizes the AAGAB:α:σ2 complex, and its binding leads to the formation of an α:σ2:CCDC32 ternary complex. The α:σ2:CCDC32 complex serves as a template that sequentially recruits the µ2 and β2 subunits of AP2 to complete AP2 assembly, accompanied by CCDC32 release. The AP2-regulating function of CCDC32 is disrupted by a disease-causing mutation. These findings demonstrate that AP2 is assembled by a handover mechanism switching from AAGAB-based initiation complexes to CCDC32-based template complexes. A similar mechanism may govern the assembly of other trafficking complexes exhibiting the same configuration as AP2.
-
WB
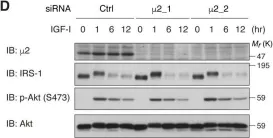
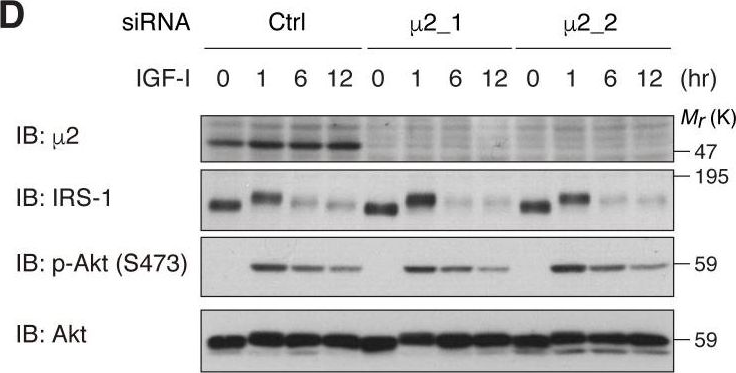
In Elife on 11 April 2018 by Yoneyama, Y., Lanzerstorfer, P., et al.
Fig.7.D
-
WB
-
Homo sapiens (Human)
Collected and cropped from eLife by CiteAb, provided under a CC-BY license
Image 1 of 3
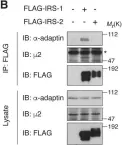
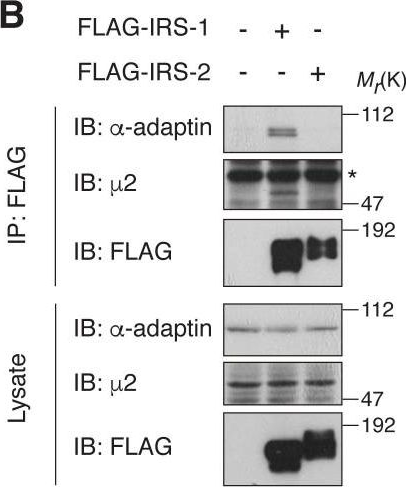
In Elife on 11 April 2018 by Yoneyama, Y., Lanzerstorfer, P., et al.
Fig.1.B
-
WB
-
Homo sapiens (Human)
Collected and cropped from eLife by CiteAb, provided under a CC-BY license
Image 1 of 3
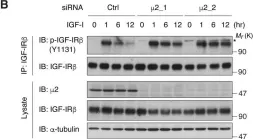
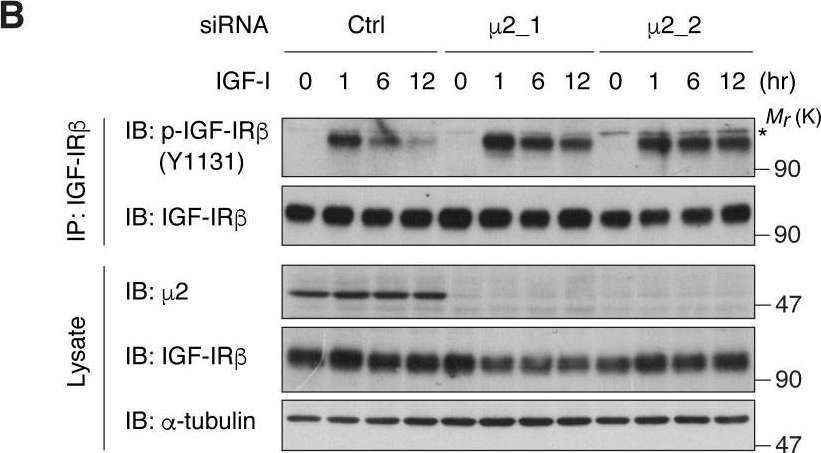
In Elife on 11 April 2018 by Yoneyama, Y., Lanzerstorfer, P., et al.
Fig.3.B
-
WB
-
Homo sapiens (Human)
Collected and cropped from eLife by CiteAb, provided under a CC-BY license
Image 1 of 3