ABSTRACT Angelman syndrome is a neurodevelopmental disorder characterized by severe motor and cognitive deficits. It is caused by the loss of the maternally inherited allele of the imprinted ubiquitin-protein ligase E3A ( UBE3A ) gene. Rodent models of Angelman syndrome do not fully recapitulate all the symptoms associated with the condition and are limited as a preclinical model for therapeutic development. Here, we show that pigs ( Sus scrofa ) with a maternally inherited deletion of UBE3A ( UBE3A −/+ ) have altered postnatal behaviors, impaired vocalizations, reduced brain growth, motor incoordination, and ataxia. Neonatal UBE3A −/+ pigs exhibited several symptoms observed in infants with Angelman syndrome, including hypotonia, suckling deficits, and failure to thrive. Collectively, these findings are consistent with the pathophysiology and developmental trajectory observed in individuals with Angelman syndrome. We anticipate that this pig model will advance our understanding of the pathophysiology of Angelman syndrome and be used as a preclinical large animal model for therapeutic development.
Product Citations: 16
Preprint on BioRxiv : the Preprint Server for Biology on 11 March 2025 by Myers, L. S., Christian, S. G., et al.
-
WB
-
Stem Cells and Developmental Biology
-
Veterinary Research
In Science Advances on 12 July 2024 by Montani, C., Balasco, L., et al.
Genomic mechanisms enhancing risk in males may contribute to sex bias in autism. The ubiquitin protein ligase E3A gene (Ube3a) affects cellular homeostasis via control of protein turnover and by acting as transcriptional coactivator with steroid hormone receptors. Overdosage of Ube3a via duplication or triplication of chromosomal region 15q11-13 causes 1 to 2% of autistic cases. Here, we test the hypothesis that increased dosage of Ube3a may influence autism-relevant phenotypes in a sex-biased manner. We show that mice with extra copies of Ube3a exhibit sex-biasing effects on brain connectomics and autism-relevant behaviors. These effects are associated with transcriptional dysregulation of autism-associated genes, as well as genes differentially expressed in 15q duplication and in autistic people. Increased Ube3a dosage also affects expression of genes on the X chromosome, genes influenced by sex steroid hormone, and genes sex-differentially regulated by transcription factors. These results suggest that Ube3a overdosage can contribute to sex bias in neurodevelopmental conditions via influence on sex-differential mechanisms.
-
WB
-
Neuroscience
In Cells on 9 September 2022 by Syding, L. A., Kubik-Zahorodna, A., et al.
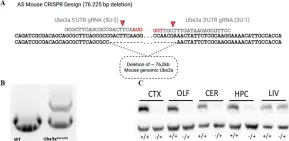
Angelman syndrome (AS) is a neurodevelopmental disorder caused by deficits in maternally inherited UBE3A. The disease is characterized by intellectual disability, impaired motor skills, and behavioral deficits, including increased anxiety and autism spectrum disorder features. The mouse models used so far in AS research recapitulate most of the cardinal AS characteristics. However, they do not mimic the situation found in the majority of AS patients who have a large deletion spanning 4-6 Mb. There is also a large variability in phenotypes reported in the available models, which altogether limits development of therapeutics. Therefore, we have generated a mouse model in which the Ube3a gene is deleted entirely from the 5' UTR to the 3' UTR of mouse Ube3a isoform 2, resulting in a deletion of 76 kb. To investigate its phenotypic suitability as a model for AS, we employed a battery of behavioral tests directed to reveal AS pathology and to find out whether this model better mirrors AS development compared to other available models. We found that the maternally inherited Ube3a-deficient line exhibits robust motor dysfunction, as seen in the rotarod and DigiGait tests, and displays abnormalities in additional behavioral paradigms, including reduced nest building and hypoactivity, although no apparent cognitive phenotype was observed in the Barnes maze and novel object recognition tests. The AS mice did, however, underperform in more complex cognition tasks, such as place reversal in the IntelliCage system, and exhibited a different circadian rhythm activity pattern. We show that the novel UBE3A-deficient model, based on a whole-gene deletion, is suitable for AS research, as it recapitulates important phenotypes characteristic of AS. This new mouse model provides complementary possibilities to study the Ube3a gene and its function in health and disease as well as possible therapeutic interventions to restore function.
-
WB
-
Mus musculus (House mouse)
-
Cell Biology
Regulation of HPV E7 Stability by E6-Associated Protein (E6AP).
In Journal of Virology on 24 August 2022 by Vats, A., Trejo-Cerro, Ó., et al.
High-risk human papillomaviruses (HPVs) are responsible for most human cervical cancers, and uncontrolled expression of the two key viral oncoproteins, E6 and E7, stimulates the induction of carcinogenesis. Previous studies have shown that both E6 and E7 are closely associated with different components of the ubiquitin proteasome pathway, including several ubiquitin ligases. Most often these are utilized to target cellular substrates for proteasome-mediated degradation, but in the case of E6, the E6AP ubiquitin ligase plays a critical role in controlling E6 stability. We now show that knockdown of E6AP in HPV-positive cervical cancer-derived cells causes a marked decrease in E7 protein levels. This is due to a decrease in the E7 half-life and occurs in a proteasome-dependent manner. In an attempt to define the underlying mechanism, we show that E7 can also associate with E6AP, albeit in a manner different from that of E6. In addition, we show that E6AP-dependent stabilization of E7 also leads to an increase in the degradation of E7's cellular target substrates. Interestingly, ectopic overexpression of E6 oncoprotein results in lower levels of E7 protein through sequestration of E6AP. We also show that increased E7 stability in the presence of E6AP increases the proliferation of the cervical cancer-derived cell lines. These results demonstrate a surprising interplay between E6 and E7, in a manner which is mediated by the E6AP ubiquitin ligase. IMPORTANCE This is the first demonstration that E6AP can directly help stabilize the HPV E7 oncoprotein, in a manner similar to that observed with HPV E6. This redefines how E6 and E7 can cooperate and potentially modulate each other's activity and further highlights the essential role played by E6AP in the viral life cycle and malignancy.
-
Homo sapiens (Human)
-
Immunology and Microbiology
In Journal of Virology on 23 March 2022 by Vats, A., Škrabar, N., et al.
Cancer-causing human papillomavirus (HPV) E6 oncoproteins contain a well-characterized phosphoacceptor site within the PDZ (PSD-95/Dlg/ZO-1) binding motif (PBM) at the C terminus of the protein. Previous studies have shown that the threonine or serine residue in the E6 PBM is subject to phosphorylation by several stress-responsive cellular kinases upon the induction of DNA damage in cervical cancer-derived cells. However, there is little information about the regulation of E6 phosphorylation in the absence of DNA damage and whether there may be other pathways by which E6 is phosphorylated. In this study, we demonstrate that loss of E6AP results in a dramatic increase in the levels of phosphorylated E6 (pE6) despite the expected overall reduction in total E6 protein levels. Furthermore, phosphorylation of E6 requires transcriptionally active p53 and occurs in a manner that is dependent upon DNA-dependent protein kinase (DNA PK). These results identify a novel feedback loop, where loss of E6AP results in upregulation of p53, leading to increased levels of E6 phosphorylation, which in turn correlates with increased association with 14-3-3 and inhibition of p53 transcriptional activity. IMPORTANCE This study demonstrates that the knockdown of E6AP from cervical cancer-derived cells leads to an increase in phosphorylation of the E6 oncoprotein. We show that this phosphorylation of E6 requires p53 transcriptional activity and the enzyme DNA PK. This study therefore defines a feedback loop whereby activation of p53 can induce phosphorylation of E6 and which in turn can inhibit p53 transcriptional activity independently of E6's ability to target p53 for degradation.
-
WB
-
Cancer Research
-
Immunology and Microbiology
In Cells on 9 September 2022 by Syding, L. A., Kubik-Zahorodna, A., et al.
Fig.1.A

-
WB
-
Mus musculus (House mouse)
Collected and cropped from Cells by CiteAb, provided under a CC-BY license
Image 1 of 1