In viable healthy cells, membrane phospholipids are asymmetrically distributed across the lipid bilayer, whereby the anionic phospholipid phosphatidylserine is virtually all distributed on the inner leaflet of the plasma membrane. During apoptosis, phospholipid asymmetry collapses and PS is externalized to the external leaflet where it serves as an “eat-me” signal for efferocytosis, the process whereby dying cells are engulfed and degraded by phagocytes. PS is also externalized on viable activated tumor endothelial cells, stromal cells and cancer cells in the tumor microenvironment reflecting a pathophysiological state of solid cancers that function to suppress host anti-tumor immunity. Several strategies have been envisioned to target dysregulated PS in the tumor microenvironment including PS binding proteins such as Annexin V and PS-targeting monoclonal antibodies (Bavituximab) with promising preclinical results. Here, in an attempt to enhance the efficacy of PS-targeting therapeutics, we have generated a series of recombinant chimeric fusion proteins that fuse type I and type III IFNs (IFN-β-IFN-λ) into a single polypeptide chain separated by a short linker. The IFN-β-IFN-λ fusion proteins retain functions of both type I and type III IFNs but show combined effects to improve biological function as well as enhance anti-tumor activities. To localize IFNs to sites of externalized PS, we next fused the IFN-β-IFN-λ chimeric protein to the PS-targeting gamma-carboxyglutamic acid-rich (Gla) domain of Growth Arrest Specific factor 6 (Gas-6), rendering these IFN biologics as PS targeting modalities. Gas6-IFN-β-IFN-λ proteins selectively bind PS as evident by solid-phase ELISA assays as well as bind PS-positive cells, including apoptotic cells and cells that express CDC50 subunit mutant of the ATP11C flippase. In vivo , Gas6-IFN-β-IFN-λ retain strong anti-tumor activities in a syngeneic model when expressed ectopically in a E0771 breast cancer model and B16-F10 melanoma models. Collectively, we report on the generation and utility of a series of novel in class IFN fusion proteins that target the immune stimulatory features of IFNs to the PS externalization in the tumor microenvironment. Abstract Figure Graphical abstract Gas6-IFN-β-IFN-λ (VitK) have tri-functional activities, acting on a diverse set of cell types to induce an anti-tumor affect. The Gas6 domain aids in homing to the PS rich tumor microenvironment, binding to apoptotic or live stressed PS positive tumor cells. The Gla domain directly binds to PS, whereas the EGF domains help in oligomerization and signal amplification resulting from intermolecular disulphide bonds. The IFN-β domain acts on immune cells such as dendritic cells and macrophages, inducing an interferon response, whereas the IFN-λ domain acts on the tumor epithelial cells, inducing tumor intrinsic anti-tumor activity.
Product Citations: 37
Preprint on BioRxiv : the Preprint Server for Biology on 26 January 2025 by Gadiyar, V., Davra, V., et al.
-
Cancer Research
The population context is a driver of the heterogeneous response of epithelial cells to interferons.
In Molecular Systems Biology on 1 March 2024 by Metz-Zumaran, C., Uckeley, Z. M., et al.
Isogenic cells respond in a heterogeneous manner to interferon. Using a micropatterning approach combined with high-content imaging and spatial analyses, we characterized how the population context (position of a cell with respect to neighboring cells) of epithelial cells affects their response to interferons. We identified that cells at the edge of cellular colonies are more responsive than cells embedded within colonies. We determined that this spatial heterogeneity in interferon response resulted from the polarized basolateral interferon receptor distribution, making cells located in the center of cellular colonies less responsive to ectopic interferon stimulation. This was conserved across cell lines and primary cells originating from epithelial tissues. Importantly, cells embedded within cellular colonies were not protected from viral infection by apical interferon treatment, demonstrating that the population context-driven heterogeneous response to interferon influences the outcome of viral infection. Our data highlights that the behavior of isolated cells does not directly translate to their behavior in a population, placing the population context as one important factor influencing heterogeneity during interferon response in epithelial cells.
© 2024. The Author(s).
-
Biochemistry and Molecular biology
Distinct Assemblies of Heterodimeric Cytokine Receptors Govern Stemness Programs in Leukemia.
In Cancer Discovery on 4 August 2023 by Kan, W. L., Dhagat, U., et al.
Leukemia stem cells (LSC) possess distinct self-renewal and arrested differentiation properties that are responsible for disease emergence, therapy failure, and recurrence in acute myeloid leukemia (AML). Despite AML displaying extensive biological and clinical heterogeneity, LSC with high interleukin-3 receptor (IL3R) levels are a constant yet puzzling feature, as this receptor lacks tyrosine kinase activity. Here, we show that the heterodimeric IL3Rα/βc receptor assembles into hexamers and dodecamers through a unique interface in the 3D structure, where high IL3Rα/βc ratios bias hexamer formation. Importantly, receptor stoichiometry is clinically relevant as it varies across the individual cells in the AML hierarchy, in which high IL3Rα/βc ratios in LSCs drive hexamer-mediated stemness programs and poor patient survival, while low ratios mediate differentiation. Our study establishes a new paradigm in which alternative cytokine receptor stoichiometries differentially regulate cell fate, a signaling mechanism that may be generalizable to other transformed cellular hierarchies and of potential therapeutic significance.
Stemness is a hallmark of many cancers and is largely responsible for disease emergence, progression, and relapse. Our finding that clinically significant stemness programs in AML are directly regulated by different stoichiometries of cytokine receptors represents a hitherto unexplained mechanism underlying cell-fate decisions in cancer stem cell hierarchies. This article is highlighted in the In This Issue feature, p. 1749.
©2023 The Authors; Published by the American Association for Cancer Research.
-
Cancer Research
Population context drives cell-to-cell variability in interferon response in epithelial cells
Preprint on BioRxiv : the Preprint Server for Biology on 26 May 2023 by Metz-Zumaran, C., Doldan, P., et al.
Isogenic cells respond in a heterogeneous manner to interferon. Using a micropatterning approach combined with high-content imaging and spatial analyses, we characterized how the population context (position of a cell with respect to the neighboring cells) of human intestinal epithelial cells affects single cell response to interferons. We identified that cells at the edge of a cellular colony are significantly more responsive than cells embedded within this colony. We determined that this spatial heterogeneity in IFN response was the result of the polarized basolateral distribution of the IFN receptors making cells located in the center of a cellular colony not responsive to ectopic IFN stimulation. We could demonstrate that this population context driven cell-to-cell variability influences the outcome of viral infection as cells embedded in a cellular colony are not protected by interferons and therefore more susceptible to infection. Our data highlights that the behavior of individual isolated cells does not directly translate to their behavior in a population, placing the population context as a key driver of cell-to-cell heterogeneity in IFN response.
-
WB
-
Homo sapiens (Human)
In Cell Reports Medicine on 19 July 2022 by Feyaerts, D., Hedou, J., et al.
The biological determinants underlying the range of coronavirus 2019 (COVID-19) clinical manifestations are not fully understood. Here, over 1,400 plasma proteins and 2,600 single-cell immune features comprising cell phenotype, endogenous signaling activity, and signaling responses to inflammatory ligands are cross-sectionally assessed in peripheral blood from 97 patients with mild, moderate, and severe COVID-19 and 40 uninfected patients. Using an integrated computational approach to analyze the combined plasma and single-cell proteomic data, we identify and independently validate a multi-variate model classifying COVID-19 severity (multi-class area under the curve [AUC]training = 0.799, p = 4.2e-6; multi-class AUCvalidation = 0.773, p = 7.7e-6). Examination of informative model features reveals biological signatures of COVID-19 severity, including the dysregulation of JAK/STAT, MAPK/mTOR, and nuclear factor κB (NF-κB) immune signaling networks in addition to recapitulating known hallmarks of COVID-19. These results provide a set of early determinants of COVID-19 severity that may point to therapeutic targets for prevention and/or treatment of COVID-19 progression.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
-
COVID-19
-
Immunology and Microbiology
In Front Immunol on 7 December 2021 by Guo, C., Reuss, D., et al.
Fig.1.A

-
WB
-
Collected and cropped from Front Immunol by CiteAb, provided under a CC-BY license
Image 1 of 12
In Front Immunol on 7 December 2021 by Guo, C., Reuss, D., et al.
Fig.1.B

-
WB
-
Collected and cropped from Front Immunol by CiteAb, provided under a CC-BY license
Image 1 of 12
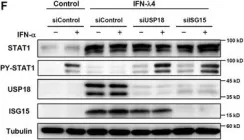
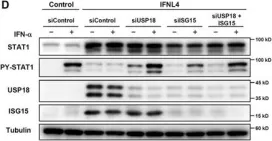
In Sci Rep on 19 June 2017 by Sung, P. S., Hong, S. H., et al.
Fig.2.B

-
WB
-
Homo sapiens (Human)
Collected and cropped from Sci Rep by CiteAb, provided under a CC-BY license
Image 1 of 12
In Sci Rep on 19 June 2017 by Sung, P. S., Hong, S. H., et al.
Fig.3.C

-
WB
-
Homo sapiens (Human)
Collected and cropped from Sci Rep by CiteAb, provided under a CC-BY license
Image 1 of 12
In Sci Rep on 19 June 2017 by Sung, P. S., Hong, S. H., et al.
Fig.3.G

-
WB
-
Homo sapiens (Human)
Collected and cropped from Sci Rep by CiteAb, provided under a CC-BY license
Image 1 of 12
In Sci Rep on 19 June 2017 by Sung, P. S., Hong, S. H., et al.
Fig.4.A

-
WB
-
Homo sapiens (Human)
Collected and cropped from Sci Rep by CiteAb, provided under a CC-BY license
Image 1 of 12
In Sci Rep on 19 June 2017 by Sung, P. S., Hong, S. H., et al.
Fig.4.C

-
WB
-
Homo sapiens (Human)
Collected and cropped from Sci Rep by CiteAb, provided under a CC-BY license
Image 1 of 12
In Sci Rep on 19 June 2017 by Sung, P. S., Hong, S. H., et al.
Fig.4.D

-
WB
-
Homo sapiens (Human)
Collected and cropped from Sci Rep by CiteAb, provided under a CC-BY license
Image 1 of 12
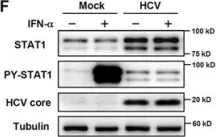
In Sci Rep on 19 June 2017 by Sung, P. S., Hong, S. H., et al.
Fig.1.F

-
WB
-
Homo sapiens (Human)
Collected and cropped from Sci Rep by CiteAb, provided under a CC-BY license
Image 1 of 12
In Sci Rep on 19 June 2017 by Sung, P. S., Hong, S. H., et al.
Fig.5.F

-
WB
-
Homo sapiens (Human)
Collected and cropped from Sci Rep by CiteAb, provided under a CC-BY license
Image 1 of 12
In Sci Rep on 19 June 2017 by Sung, P. S., Hong, S. H., et al.
Fig.5.D

-
WB
-
Homo sapiens (Human)
Collected and cropped from Sci Rep by CiteAb, provided under a CC-BY license
Image 1 of 12
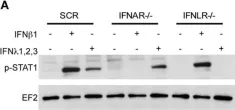
In Front Immunol on 10 May 2017 by Pervolaraki, K., Stanifer, M. L., et al.
Fig.6.A

-
WB
-
Homo sapiens (Human)
Collected and cropped from Front Immunol by CiteAb, provided under a CC-BY license
Image 1 of 12